Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1
- PMID: 23055555
- PMCID: PMC3503140
- DOI: 10.1128/JVI.01998-12
Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1
Abstract
HLA class I-associated polymorphisms identified at the population level mark viral sites under immune pressure by individual HLA alleles. As such, analysis of their distribution, frequency, location, statistical strength, sequence conservation, and other properties offers a unique perspective from which to identify correlates of protective cellular immunity. We analyzed HLA-associated HIV-1 subtype B polymorphisms in 1,888 treatment-naïve, chronically infected individuals using phylogenetically informed methods and identified characteristics of HLA-associated immune pressures that differentiate protective and nonprotective alleles. Over 2,100 HLA-associated HIV-1 polymorphisms were identified, approximately one-third of which occurred inside or within 3 residues of an optimally defined cytotoxic T-lymphocyte (CTL) epitope. Differential CTL escape patterns between closely related HLA alleles were common and increased with greater evolutionary distance between allele group members. Among 9-mer epitopes, mutations at HLA-specific anchor residues represented the most frequently detected escape type: these occurred nearly 2-fold more frequently than expected by chance and were computationally predicted to reduce peptide-HLA binding nearly 10-fold on average. Characteristics associated with protective HLA alleles (defined using hazard ratios for progression to AIDS from natural history cohorts) included the potential to mount broad immune selection pressures across all HIV-1 proteins except Nef, the tendency to drive multisite and/or anchor residue escape mutations within known CTL epitopes, and the ability to strongly select mutations in conserved regions within HIV's structural and functional proteins. Thus, the factors defining protective cellular immune responses may be more complex than simply targeting conserved viral regions. The results provide new information to guide vaccine design and immunogenicity studies.
Figures
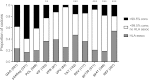
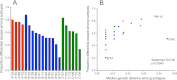

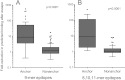

References
-
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19:716–723
Publication types
MeSH terms
Substances
Grants and funding
- 1U01-AI069484/AI/NIAID NIH HHS/United States
- U01 AI069502/AI/NIAID NIH HHS/United States
- U01 AI069513/AI/NIAID NIH HHS/United States
- AI 69467-01/AI/NIAID NIH HHS/United States
- UM1 AI069494/AI/NIAID NIH HHS/United States
- UM1 AI069423/AI/NIAID NIH HHS/United States
- AI38858-09S1/AI/NIAID NIH HHS/United States
- U01 AI069474/AI/NIAID NIH HHS/United States
- AI32782/AI/NIAID NIH HHS/United States
- U01 AI069434/AI/NIAID NIH HHS/United States
- AI-69465/AI/NIAID NIH HHS/United States
- AI069556/AI/NIAID NIH HHS/United States
- U01 AI069447/AI/NIAID NIH HHS/United States
- UM1 AI069501/AI/NIAID NIH HHS/United States
- UM1 AI069472/AI/NIAID NIH HHS/United States
- U01 AI069467/AI/NIAID NIH HHS/United States
- U01 AI069423/AI/NIAID NIH HHS/United States
- UM1 AI069513/AI/NIAID NIH HHS/United States
- M01 RR000096/RR/NCRR NIH HHS/United States
- M01 RR000046/RR/NCRR NIH HHS/United States
- AI46370/AI/NIAID NIH HHS/United States
- U01 AI027661/AI/NIAID NIH HHS/United States
- UM1 AI069424/AI/NIAID NIH HHS/United States
- AI69472/AI/NIAID NIH HHS/United States
- AI-069439/AI/NIAID NIH HHS/United States
- U01 AI069465/AI/NIAID NIH HHS/United States
- UM1 AI069434/AI/NIAID NIH HHS/United States
- UL1 RR024996/RR/NCRR NIH HHS/United States
- K24 AI064086/AI/NIAID NIH HHS/United States
- UM1 AI069411/AI/NIAID NIH HHS/United States
- 1 U01 AI069452-01/AI/NIAID NIH HHS/United States
- UM1 AI069432/AI/NIAID NIH HHS/United States
- AI069532/AI/NIAID NIH HHS/United States
- AI046376-05/AI/NIAID NIH HHS/United States
- AI069434/AI/NIAID NIH HHS/United States
- M01 RR-00032/RR/NCRR NIH HHS/United States
- AI069474/AI/NIAID NIH HHS/United States
- UM1 AI069495/AI/NIAID NIH HHS/United States
- U01 AI069484/AI/NIAID NIH HHS/United States
- AI36214/AI/NIAID NIH HHS/United States
- UM1 AI069471/AI/NIAID NIH HHS/United States
- U01 AI069439/AI/NIAID NIH HHS/United States
- U01 AI069556/AI/NIAID NIH HHS/United States
- AI 069471/AI/NIAID NIH HHS/United States
- M01-RR00096/RR/NCRR NIH HHS/United States
- RR00051/RR/NCRR NIH HHS/United States
- U01 AI027670/AI/NIAID NIH HHS/United States
- TL1 RR024978/RR/NCRR NIH HHS/United States
- KL2 RR024977/RR/NCRR NIH HHS/United States
- AI 069501/AI/NIAID NIH HHS/United States
- U01 AI069532/AI/NIAID NIH HHS/United States
- UM1 AI069439/AI/NIAID NIH HHS/United States
- M01 RR002635/RR/NCRR NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- M01 RR000044/RR/NCRR NIH HHS/United States
- U01 AI069501/AI/NIAID NIH HHS/United States
- AI69411/AI/NIAID NIH HHS/United States
- AI34853/AI/NIAID NIH HHS/United States
- M01 RR000750/RR/NCRR NIH HHS/United States
- U01 AI046376/AI/NIAID NIH HHS/United States
- MO1RR000750/RR/NCRR NIH HHS/United States
- AI069495/AI/NIAID NIH HHS/United States
- MOP-93536/CAPMC/ CIHR/Canada
- UM1 AI069477/AI/NIAID NIH HHS/United States
- U01 AI069432/AI/NIAID NIH HHS/United States
- AI27670/AI/NIAID NIH HHS/United States
- AI068636/AI/NIAID NIH HHS/United States
- U01 AI069450/AI/NIAID NIH HHS/United States
- U01 AI038858/AI/NIAID NIH HHS/United States
- RR024975/RR/NCRR NIH HHS/United States
- AI69494-01/AI/NIAID NIH HHS/United States
- RR02635/RR/NCRR NIH HHS/United States
- U01 AI046370/AI/NIAID NIH HHS/United States
- HOP-115700/CAPMC/ CIHR/Canada
- U01 AI068636/AI/NIAID NIH HHS/United States
- KL2 TR000458/TR/NCATS NIH HHS/United States
- UM1 AI069474/AI/NIAID NIH HHS/United States
- AI060354/AI/NIAID NIH HHS/United States
- M01 RR000052/RR/NCRR NIH HHS/United States
- U01 AI034853/AI/NIAID NIH HHS/United States
- AI069419-01/AI/NIAID NIH HHS/United States
- AI069424/AI/NIAID NIH HHS/United States
- R01 AI060460/AI/NIAID NIH HHS/United States
- M01 RR000051/RR/NCRR NIH HHS/United States
- U01 AI069495/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- UM1 AI069556/AI/NIAID NIH HHS/United States
- P30 AI110527/AI/NIAID NIH HHS/United States
- M01 RR000032/RR/NCRR NIH HHS/United States
- U01 AI027673/AI/NIAID NIH HHS/United States
- AI069502-01/AI/NIAID NIH HHS/United States
- AI 27661/AI/NIAID NIH HHS/United States
- AI064086/AI/NIAID NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- UM1 AI069502/AI/NIAID NIH HHS/United States
- U01 AI025859/AI/NIAID NIH HHS/United States
- UM1 AI069450/AI/NIAID NIH HHS/United States
- UM1 AI069532/AI/NIAID NIH HHS/United States
- AI27673/AI/NIAID NIH HHS/United States
- UM1 AI069465/AI/NIAID NIH HHS/United States
- U01 AI069424/AI/NIAID NIH HHS/United States
- U01 AI032782/AI/NIAID NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- AI-069513/AI/NIAID NIH HHS/United States
- AI069450/AI/NIAID NIH HHS/United States
- RR-00052/RR/NCRR NIH HHS/United States
- U01 AI069411/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- AI069472/AI/NIAID NIH HHS/United States
- 5-P30-AI-045008-07/AI/NIAID NIH HHS/United States
- UM1 AI069419/AI/NIAID NIH HHS/United States
- AI069432/AI/NIAID NIH HHS/United States
- U01 AI069477/AI/NIAID NIH HHS/United States
- 5-MO1 RR00044/RR/NCRR NIH HHS/United States
- AI50410/AI/NIAID NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
- U01 AI069419/AI/NIAID NIH HHS/United States
- AI25859/AI/NIAID NIH HHS/United States
- UL1 TR000457/TR/NCATS NIH HHS/United States
- U01 AI069494/AI/NIAID NIH HHS/United States
- UM1 AI069447/AI/NIAID NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- U01 AI069452/AI/NIAID NIH HHS/United States
- UM1 AI069467/AI/NIAID NIH HHS/United States
- AI69423-01/AI/NIAID NIH HHS/United States
- AI069447-01/AI/NIAID NIH HHS/United States
- AI069477/AI/NIAID NIH HHS/United States
- RR024996/RR/NCRR NIH HHS/United States
- RR00046/RR/NCRR NIH HHS/United States
- U01 AI069471/AI/NIAID NIH HHS/United States
- UOIA138858/PHS HHS/United States
- U01 AI069472/AI/NIAID NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

