Diversity and distribution of hantaviruses in South America
- PMID: 23055565
- PMCID: PMC3503106
- DOI: 10.1128/JVI.02341-12
Diversity and distribution of hantaviruses in South America
Abstract
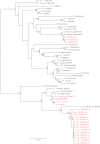
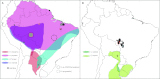
Hantaviruses are important contributors to disease burden in the New World, yet many aspects of their distribution and dynamics remain uncharacterized. To examine the patterns and processes that influence the diversity and geographic distribution of hantaviruses in South America, we performed genetic and phylogeographic analyses of all available South American hantavirus sequences. We sequenced multiple novel and previously described viruses (Anajatuba, Laguna Negra-like, two genotypes of Castelo dos Sonhos, and two genotypes of Rio Mamore) from Brazilian Oligoryzomys rodents and hantavirus pulmonary syndrome cases and identified a previously uncharacterized species of Oligoryzomys associated with a new genotype of Rio Mamore virus. Our analysis indicates that the majority of South American hantaviruses fall into three phylogenetic clades, corresponding to Andes and Andes-like viruses, Laguna Negra and Laguna Negra-like viruses, and Rio Mamore and Rio Mamore-like viruses. In addition, the dynamics and distribution of these viruses appear to be shaped by both the geographic proximity and phylogenetic relatedness of their rodent hosts. The current system of nomenclature used in the hantavirus community is a significant impediment to understanding the ecology and evolutionary history of hantaviruses; here, we suggest strict adherence to a modified taxonomic system, with species and strain designations resembling the numerical system of the enterovirus genus.
Figures



References
-
- Baele G, et al. 7 March 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. [Epub ahead of print.] doi:10.1093/molbev/mss084 - DOI - PMC - PubMed
-
- Bharadwaj M, Botten J, Torrez-Martinez N, Hjelle B. 1997. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am. J. Trop. Med. Hyg. 57:368–374 - PubMed
-
- Campos GM, et al. 2003. Serological survey of hantavirus in Jardinopolis County, Brazil. J. Med. Virol. 71:417–422 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

