Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope
- PMID: 23055922
- PMCID: PMC3464233
- DOI: 10.1371/journal.ppat.1002930
Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope
Abstract
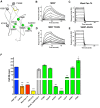
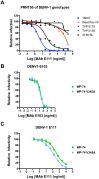
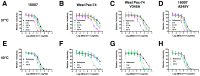
We previously developed a panel of neutralizing monoclonal antibodies against Dengue virus (DENV)-1, of which few exhibited inhibitory activity against all DENV-1 genotypes. This finding is consistent with reports observing variable neutralization of different DENV strains and genotypes using serum from individuals that experienced natural infection or immunization. Herein, we describe the crystal structures of DENV1-E111 bound to a novel CC' loop epitope on domain III (DIII) of the E protein from two different DENV-1 genotypes. Docking of our structure onto the available cryo-electron microscopy models of DENV virions revealed that the DENV1-E111 epitope was inaccessible, suggesting that this antibody recognizes an uncharacterized virus conformation. While the affinity of binding between DENV1-E111 and DIII varied by genotype, we observed limited correlation with inhibitory activity. Instead, our results support the conclusion that potent neutralization depends on genotype-dependent exposure of the CC' loop epitope. These findings establish new structural complexity of the DENV virion, which may be relevant for the choice of DENV strain for induction or analysis of neutralizing antibodies in the context of vaccine development.
Conflict of interest statement
MacroGenics has licensed some of the anti-DENV antibodies described in the work from Washington University. Syd Johnson is an employee of MacroGenics. Michael Diamond is a paid consultant of MacroGenics. None of the other authors have financial interests related to this work. This does not alter our adherence to all PLOS Pathogens policies on sharing data and materials.
Figures



References
-
- WHO. Dengue and severe dengue. Available: http://www.who.int/mediacentre/factsheets/fs117/en/index.html. Accessed 27 May 2012.
-
- Rico-Hesse R (1990) Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174: 479–493. - PubMed
-
- Holmes EC, Twiddy SS (2003) The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol 3: 19–28. - PubMed
-
- Sabin AB (1952) Research on Dengue during World War II. Am J Trop Med Hyg 1: 30–50. - PubMed
-
- Halstead SB, Nimmannitya S, Yamarat C, Russell PK (1967) Hemorrhagic fever in Thailand; recent knowledge regarding etiology. Jpn J Med Sci Biol 20: 96–103. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

