A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse
- PMID: 23055939
- PMCID: PMC3464207
- DOI: 10.1371/journal.pgen.1002966
A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse
Abstract
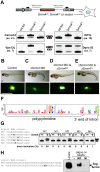
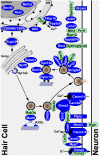
Sensory hair cells are essential for hearing and balance. Their development from epithelial precursors has been extensively characterized with respect to transcriptional regulation, but not in terms of posttranscriptional influences. Here we report on the identification and functional characterization of an alternative-splicing regulator whose inactivation is responsible for defective hair-cell development, deafness, and impaired balance in the spontaneous mutant Bronx waltzer (bv) mouse. We used positional cloning and transgenic rescue to locate the bv mutation to the splicing factor-encoding gene Ser/Arg repetitive matrix 4 (Srrm4). Transcriptome-wide analysis of pre-mRNA splicing in the sensory patches of embryonic inner ears revealed that specific alternative exons were skipped at abnormally high rates in the bv mice. Minigene experiments in a heterologous expression system confirmed that these skipped exons require Srrm4 for inclusion into the mature mRNA. Sequence analysis and mutagenesis experiments showed that the affected transcripts share a novel motif that is necessary for the Srrm4-dependent alternative splicing. Functional annotations and protein-protein interaction data indicated that the encoded proteins cluster in the secretion and neurotransmission pathways. In addition, the splicing of a few transcriptional regulators was found to be Srrm4 dependent, and several of the genes known to be targeted by these regulators were expressed at reduced levels in the bv mice. Although Srrm4 expression was detected in neural tissues as well as hair cells, analyses of the bv mouse cerebellum and neocortex failed to detect splicing defects. Our data suggest that Srrm4 function is critical in the hearing and balance organs, but not in all neural tissues. Srrm4 is the first alternative-splicing regulator to be associated with hearing, and the analysis of bv mice provides exon-level insights into hair-cell development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
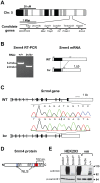
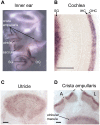
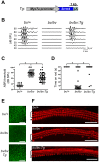
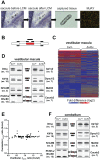
References
-
- Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ (2004) Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet 5: 489–498. - PubMed
-
- Richardson GP, de Monvel JB, Petit C (2011) How the genetics of deafness illuminates auditory physiology. Annu Rev Physiol 73: 311–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

