Improving PacBio long read accuracy by short read alignment
- PMID: 23056399
- PMCID: PMC3464235
- DOI: 10.1371/journal.pone.0046679
Improving PacBio long read accuracy by short read alignment
Abstract
The recent development of third generation sequencing (TGS) generates much longer reads than second generation sequencing (SGS) and thus provides a chance to solve problems that are difficult to study through SGS alone. However, higher raw read error rates are an intrinsic drawback in most TGS technologies. Here we present a computational method, LSC, to perform error correction of TGS long reads (LR) by SGS short reads (SR). Aiming to reduce the error rate in homopolymer runs in the main TGS platform, the PacBio® RS, LSC applies a homopolymer compression (HC) transformation strategy to increase the sensitivity of SR-LR alignment without scarifying alignment accuracy. We applied LSC to 100,000 PacBio long reads from human brain cerebellum RNA-seq data and 64 million single-end 75 bp reads from human brain RNA-seq data. The results show LSC can correct PacBio long reads to reduce the error rate by more than 3 folds. The improved accuracy greatly benefits many downstream analyses, such as directional gene isoform detection in RNA-seq study. Compared with another hybrid correction tool, LSC can achieve over double the sensitivity and similar specificity.
Conflict of interest statement
Figures
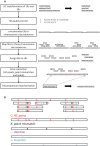
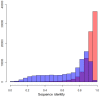
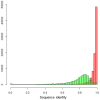
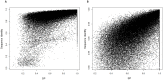
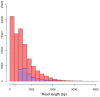
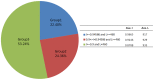

References
-
- HiSeq™ Sequencing Systems - Redefining the trajectory of sequencing. Available: http://www.illumina.com/Documents/systems/hiseq/datasheet_hiseq_systems.pdf. Accessed 2012 Sep 8.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

