Dynamic reorganization of metabolic enzymes into intracellular bodies
- PMID: 23057741
- PMCID: PMC4089986
- DOI: 10.1146/annurev-cellbio-101011-155841
Dynamic reorganization of metabolic enzymes into intracellular bodies
Abstract
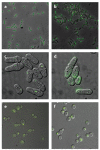
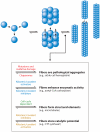
Both focused and large-scale cell biological and biochemical studies have revealed that hundreds of metabolic enzymes across diverse organisms form large intracellular bodies. These proteinaceous bodies range in form from fibers and intracellular foci--such as those formed by enzymes of nitrogen and carbon utilization and of nucleotide biosynthesis--to high-density packings inside bacterial microcompartments and eukaryotic microbodies. Although many enzymes clearly form functional mega-assemblies, it is not yet clear for many recently discovered cases whether they represent functional entities, storage bodies, or aggregates. In this article, we survey intracellular protein bodies formed by metabolic enzymes, asking when and why such bodies form and what their formation implies for the functionality--and dysfunctionality--of the enzymes that comprise them. The panoply of intracellular protein bodies also raises interesting questions regarding their evolution and maintenance within cells. We speculate on models for how such structures form in the first place and why they may be inevitable.
Figures





References
-
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

