The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na(+) loading in xylem and confers salt tolerance in transgenic tobacco
- PMID: 23057782
- PMCID: PMC3548769
- DOI: 10.1186/1471-2229-12-188
The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na(+) loading in xylem and confers salt tolerance in transgenic tobacco
Abstract
Background: Soil salinity adversely affects plant growth and development and disturbs intracellular ion homeostasis resulting cellular toxicity. The Salt Overly Sensitive 1 (SOS1) gene encodes a plasma membrane Na(+)/H(+) antiporter that plays an important role in imparting salt stress tolerance to plants. Here, we report the cloning and characterisation of the SbSOS1 gene from Salicornia brachiata, an extreme halophyte.
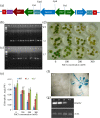
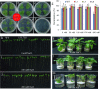
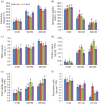
Results: The SbSOS1 gene is 3774 bp long and encodes a protein of 1159 amino acids. SbSOS1 exhibited a greater level of constitutive expression in roots than in shoots and was further increased by salt stress. Overexpressing the S. brachiata SbSOS1 gene in tobacco conferred high salt tolerance, promoted seed germination and increased root length, shoot length, leaf area, fresh weight, dry weight, relative water content (RWC), chlorophyll, K(+)/Na(+) ratio, membrane stability index, soluble sugar, proline and amino acid content relative to wild type (WT) plants. Transgenic plants exhibited reductions in electrolyte leakage, reactive oxygen species (ROS) and MDA content in response to salt stress, which probably occurred because of reduced cytosolic Na(+) content and oxidative damage. At higher salt stress, transgenic tobacco plants exhibited reduced Na(+) content in root and leaf and higher concentrations in stem and xylem sap relative to WT, which suggests a role of SbSOS1 in Na(+) loading to xylem from root and leaf tissues. Transgenic lines also showed increased K(+) and Ca(2+) content in root tissue compared to WT, which reflect that SbSOS1 indirectly affects the other transporters activity.
Conclusions: Overexpression of SbSOS1 in tobacco conferred a high degree of salt tolerance, enhanced plant growth and altered physiological and biochemical parameters in response to salt stress. In addition to Na(+) efflux outside the plasma membrane, SbSOS1 also helps to maintain variable Na(+) content in different organs and also affect the other transporters activity indirectly. These results broaden the role of SbSOS1 in planta and suggest that this gene could be used to develop salt-tolerant transgenic crops.
Figures

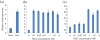



References
-
- Epstein E, Bloom AJ. Mineral nutrition of plants: principles and perspectives. 2. Sunderland, Massachusetts: Sinauer Associations; 2005. pp. 342–351.
-
- FAO. FAO land and plant nutrition management service. 2008. http://www.fao.org/ag/agl/agll/spush/
-
- Wyn Jones RG, Pollard A. Encyclopedia of plant physiology. Berlin: Springer- Verlag; 1983.
-
- Hernández JA, Olmos E, Corpas FJ, Sevilla F, Del Río LA. Salt-induced oxidative stress in chloroplasts of pea plants. Plant Sci. 1995;105:151–167. doi: 10.1016/0168-9452(94)04047-8. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

