Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene
- PMID: 23057995
- PMCID: PMC3548740
- DOI: 10.1186/1471-2229-12-190
Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene
Abstract
Background: The phytohormone ethylene is involved in a wide range of developmental processes and in mediating plant responses to biotic and abiotic stresses. Ethylene signalling acts via a linear transduction pathway leading to the activation of Ethylene Response Factor genes (ERF) which represent one of the largest gene families of plant transcription factors. How an apparently simple signalling pathway can account for the complex and widely diverse plant responses to ethylene remains yet an unanswered question. Building on the recent release of the complete tomato genome sequence, the present study aims at gaining better insight on distinctive features among ERF proteins.
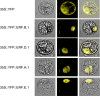
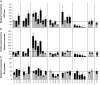
Results: A set of 28 cDNA clones encoding ERFs in the tomato (Solanum lycopersicon) were isolated and shown to fall into nine distinct subclasses characterised by specific conserved motifs most of which with unknown function. In addition of being able to regulate the transcriptional activity of GCC-box containing promoters, tomato ERFs are also shown to be active on promoters lacking this canonical ethylene-responsive-element. Moreover, the data reveal that ERF affinity to the GCC-box depends on the nucleotide environment surrounding this cis-acting element. Site-directed mutagenesis revealed that the nature of the flanking nucleotides can either enhance or reduce the binding affinity, thus conferring the binding specificity of various ERFs to target promoters.Based on their expression pattern, ERF genes can be clustered in two main clades given their preferential expression in reproductive or vegetative tissues. The regulation of several tomato ERF genes by both ethylene and auxin, suggests their potential contribution to the convergence mechanism between the signalling pathways of the two hormones.
Conclusions: The data reveal that regions flanking the core GCC-box sequence are part of the discrimination mechanism by which ERFs selectively bind to their target promoters. ERF tissue-specific expression combined to their responsiveness to both ethylene and auxin bring some insight on the complexity and fine regulation mechanisms involving these transcriptional mediators. All together the data support the hypothesis that ERFs are the main component enabling ethylene to regulate a wide range of physiological processes in a highly specific and coordinated manner.
Figures




References
-
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. - PubMed
-
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–254. - PubMed
-
- Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, Regad F, Latché A, Pech JC, Bouzayen M. Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol. 2006;47:1195–1205. - PubMed
-
- Benavente LM, Alonso JM. Molecular mechanisms of ethylene signaling in Arabidopsis. Mol Biosyst. 2006;2:165–173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

