HIF-1α downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation
- PMID: 23059786
- PMCID: PMC3569981
- DOI: 10.1038/cdd.2012.130
HIF-1α downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation
Abstract
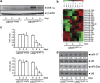
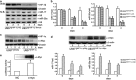
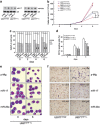
Hypoxia-inducible factor 1 (HIF-1) is a crucial transcription factor for the cellular adaptive response to hypoxia, which contributes to multiple events in cancer biology. MicroRNAs (miRNAs) are involved in almost all cellular activities such as differentiation, proliferation, and apoptosis. In this work, we use miRNA microarrays to profile miRNA expression in acute myeloid leukemia (AML) cells with inducible HIF-1α expression, and identify 19 differentially expressed miRNAs. Our study shows that HIF-1α represses the expression of miR-17 and miR-20a by downregulating c-Myc expression. These two miRNAs alleviate hypoxia and HIF-1α-induced differentiation of AML cells. More intriguingly, miR-17 and miR-20a directly inhibit the p21 and STAT3 (signal transducer and activator of transcription 3) expression, both of which can reverse miR-17/miR-20a-mediated abrogation of HIF-1α-induced differentiation. Moreover, we show in vivo that miR-20a contributes to HIF-1α-induced differentiation of leukemic cells. Taken together, our results suggest that HIF-1α regulates the miRNA network to interfere with AML cell differentiation, representing a novel molecular mechanism for HIF-1-mediated anti-leukemic action.
Figures


References
-
- Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 365:537–547. - PubMed
-
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. - PubMed
-
- Nicholas SA, Sumbayev VV. The involvement of hypoxia-inducible factor 1 alpha in Toll-like receptor 7/8-mediated inflammatory response. Cell Res. 2009;19:973–983. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Huang Y, Du KM, Xue ZH, Yan H, Li D, Liu W, et al. Cobalt chloride and low oxygen tension trigger differentiation of acute myeloid leukemic cells: possible mediation of hypoxia-inducible factor-1alpha. Leukemia. 2003;17:2065–2073. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

