Network-based inference from complex proteomic mixtures using SNIPE
- PMID: 23060611
- PMCID: PMC3509492
- DOI: 10.1093/bioinformatics/bts594
Network-based inference from complex proteomic mixtures using SNIPE
Abstract
Motivation: Proteomics presents the opportunity to provide novel insights about the global biochemical state of a tissue. However, a significant problem with current methods is that shotgun proteomics has limited success at detecting many low abundance proteins, such as transcription factors from complex mixtures of cells and tissues. The ability to assay for these proteins in the context of the entire proteome would be useful in many areas of experimental biology.
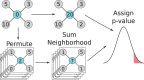
Results: We used network-based inference in an approach named SNIPE (Software for Network Inference of Proteomics Experiments) that selectively highlights proteins that are more likely to be active but are otherwise undetectable in a shotgun proteomic sample. SNIPE integrates spectral counts from paired case-control samples over a network neighbourhood and assesses the statistical likelihood of enrichment by a permutation test. As an initial application, SNIPE was able to select several proteins required for early murine tooth development. Multiple lines of additional experimental evidence confirm that SNIPE can uncover previously unreported transcription factors in this system. We conclude that SNIPE can enhance the utility of shotgun proteomics data to facilitate the study of poorly detected proteins in complex mixtures.
Availability and implementation: An implementation for the R statistical computing environment named snipeR has been made freely available at http://genetics.bwh.harvard.edu/snipe/.
Contact: ssunyaev@rics.bwh.harvard.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures



References
-
- Bantscheff M, et al. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. - PubMed
-
- Csárdi G, Nepusz T. The igraph software package for complex network research. Inter J. Complex Syst. 2006;1695
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

