Gene expression profiling following NRF2 and KEAP1 siRNA knockdown in human lung fibroblasts identifies CCL11/Eotaxin-1 as a novel NRF2 regulated gene
- PMID: 23061798
- PMCID: PMC3546844
- DOI: 10.1186/1465-9921-13-92
Gene expression profiling following NRF2 and KEAP1 siRNA knockdown in human lung fibroblasts identifies CCL11/Eotaxin-1 as a novel NRF2 regulated gene
Abstract
Background: Oxidative Stress contributes to the pathogenesis of many diseases. The NRF2/KEAP1 axis is a key transcriptional regulator of the anti-oxidant response in cells. Nrf2 knockout mice have implicated this pathway in regulating inflammatory airway diseases such as asthma and COPD. To better understand the role the NRF2 pathway has on respiratory disease we have taken a novel approach to define NRF2 dependent gene expression in a relevant lung system.
Methods: Normal human lung fibroblasts were transfected with siRNA specific for NRF2 or KEAP1. Gene expression changes were measured at 30 and 48 hours using a custom Affymetrix Gene array. Changes in Eotaxin-1 gene expression and protein secretion were further measured under various inflammatory conditions with siRNAs and pharmacological tools.
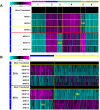
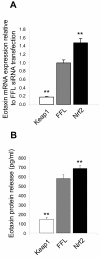
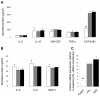
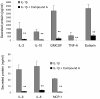
Results: An anti-correlated gene set (inversely regulated by NRF2 and KEAP1 RNAi) that reflects specific NRF2 regulated genes was identified. Gene annotations show that NRF2-mediated oxidative stress response is the most significantly regulated pathway, followed by heme metabolism, metabolism of xenobiotics by Cytochrome P450 and O-glycan biosynthesis. Unexpectedly the key eosinophil chemokine Eotaxin-1/CCL11 was found to be up-regulated when NRF2 was inhibited and down-regulated when KEAP1 was inhibited. This transcriptional regulation leads to modulation of Eotaxin-1 secretion from human lung fibroblasts under basal and inflammatory conditions, and is specific to Eotaxin-1 as NRF2 or KEAP1 knockdown had no effect on the secretion of a set of other chemokines and cytokines. Furthermore, the known NRF2 small molecule activators CDDO and Sulphoraphane can also dose dependently inhibit Eotaxin-1 release from human lung fibroblasts.
Conclusions: These data uncover a previously unknown role for NRF2 in regulating Eotaxin-1 expression and further the mechanistic understanding of this pathway in modulating inflammatory lung disease.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

