Retention of conformational entropy upon calmodulin binding to target peptides is driven by transient salt bridges
- PMID: 23062350
- PMCID: PMC3471477
- DOI: 10.1016/j.bpj.2012.08.037
Retention of conformational entropy upon calmodulin binding to target peptides is driven by transient salt bridges
Abstract
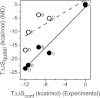
Calmodulin (CaM) is a highly flexible calcium-binding protein that mediates signal transduction through an ability to differentially bind to highly variable binding sequences in target proteins. To identify how binding affects CaM motions, and its relationship to conformational entropy and target peptide sequence, we have employed fully atomistic, explicit solvent molecular dynamics simulations of unbound CaM and CaM bound to five different target peptides. The calculated CaM conformational binding entropies correlate with experimentally derived conformational entropies with a correlation coefficient R(2) of 0.95. Selected side-chain interactions with target peptides restrain interhelical loop motions, acting to tune the conformational entropy of the bound complex via widely distributed CaM motions. In the complex with the most conformational entropy retention (CaM in complex with the neuronal nitric oxide synthase binding sequence), Lys-148 at the C-terminus of CaM forms transient salt bridges alternating between Glu side chains in the N-domain, the central linker, and the binding target. Additional analyses of CaM structures, fluctuations, and CaM-target interactions illuminate the interplay between electrostatic, side chain, and backbone properties in the ability of CaM to recognize and discriminate against targets by tuning its conformational entropy, and suggest a need to consider conformational dynamics in optimizing binding affinities.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Chin D., Means A.R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. - PubMed
-
- Afshar M., Caves L.S.D., Haiech J. Investigating the high affinity and low sequence specificity of calmodulin binding to its targets. J. Mol. Biol. 1994;244:554–571. - PubMed
-
- Grünberg R., Nilges M., Leckner J. Flexibility and conformational entropy in protein-protein binding. Structure. 2006;14:683–693. - PubMed
-
- Brady G.P., Sharp K.A. Entropy in protein folding and in protein-protein interactions. Curr. Opin. Struct. Biol. 1997;7:215–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

