Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome
- PMID: 23063120
- PMCID: PMC3490196
- DOI: 10.1016/j.cell.2012.09.024
Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome
Abstract
Pathogenic simian immunodeficiency virus (SIV) infection is associated with enteropathy, which likely contributes to AIDS progression. To identify candidate etiologies for AIDS enteropathy, we used next-generation sequencing to define the enteric virome during SIV infection in nonhuman primates. Pathogenic, but not nonpathogenic, SIV infection was associated with significant expansion of the enteric virome. We identified at least 32 previously undescribed enteric viruses during pathogenic SIV infection and confirmed their presence by using viral culture and PCR testing. We detected unsuspected mucosal adenovirus infection associated with enteritis as well as parvovirus viremia in animals with advanced AIDS, indicating the pathogenic potential of SIV-associated expansion of the enteric virome. No association between pathogenic SIV infection and the family-level taxonomy of enteric bacteria was detected. Thus, enteric viral infections may contribute to AIDS enteropathy and disease progression. These findings underline the importance of metagenomic analysis of the virome for understanding AIDS pathogenesis.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
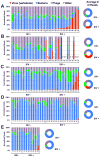


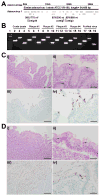

Comment in
-
Viral pathogenesis: SIV gives the virome a boost.Nat Rev Microbiol. 2012 Dec;10(12):805. doi: 10.1038/nrmicro2919. Epub 2012 Nov 13. Nat Rev Microbiol. 2012. PMID: 23147710 No abstract available.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. JMolBiol. 1990;215:403–410. - PubMed
-
- Bailey C, Mansfield K. Emerging and reemerging infectious diseases of nonhuman primates in the laboratory setting. VetPathol. 2010;47:462–481. - PubMed
-
- Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- U54 AI057160-08/AI/NIAID NIH HHS/United States
- U19 AI078526/AI/NIAID NIH HHS/United States
- AI066305/AI/NIAID NIH HHS/United States
- R01 RR032309/RR/NCRR NIH HHS/United States
- R01 AI066924/AI/NIAID NIH HHS/United States
- P51 OD011103/OD/NIH HHS/United States
- U19 AI095985/AI/NIAID NIH HHS/United States
- AI078526/AI/NIAID NIH HHS/United States
- AI65335/AI/NIAID NIH HHS/United States
- AI066924/AI/NIAID NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
- 8P51OD011103-5/OD/NIH HHS/United States
- R01 OD011170/OD/NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
- U19 AI066305/AI/NIAID NIH HHS/United States
- R01 AI065335/AI/NIAID NIH HHS/United States
- AI095985/AI/NIAID NIH HHS/United States
- OD11170-02/OD/NIH HHS/United States
- K08 AI087992/AI/NIAID NIH HHS/United States
- 1R01 RR032309/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

