A resource for manipulating gene expression and analyzing cis-regulatory modules in the Drosophila CNS
- PMID: 23063363
- PMCID: PMC3523218
- DOI: 10.1016/j.celrep.2012.09.009
A resource for manipulating gene expression and analyzing cis-regulatory modules in the Drosophila CNS
Abstract
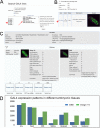
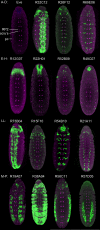
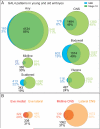
Here, we describe the embryonic central nervous system expression of 5,000 GAL4 lines made using molecularly defined cis-regulatory DNA inserted into a single attP genomic location. We document and annotate the patterns in early embryos when neurogenesis is at its peak, and in older embryos where there is maximal neuronal diversity and the first neural circuits are established. We note expression in other tissues, such as the lateral body wall (muscle, sensory neurons, and trachea) and viscera. Companion papers report on the adult brain and larval imaginal discs, and the integrated data sets are available online (http://www.janelia.org/gal4-gen1). This collection of embryonically expressed GAL4 lines will be valuable for determining neuronal morphology and function. The 1,862 lines expressed in small subsets of neurons (<20/segment) will be especially valuable for characterizing interneuronal diversity and function, because although interneurons comprise the majority of all central nervous system neurons, their gene expression profile and function remain virtually unexplored.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Akiyama-Oda Y, Hosoya T, Hotta Y. Asymmetric cell division of thoracic neuroblast 6–4 to bifurcate glial and neuronal lineage in Drosophila. Development. 1999;126:1967–1974. - PubMed
-
- Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128:4251–4264. - PubMed
-
- Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

