Advances in ion mobility spectrometry-mass spectrometry reveal key insights into amyloid assembly
- PMID: 23063533
- PMCID: PMC3787735
- DOI: 10.1016/j.bbapap.2012.10.002
Advances in ion mobility spectrometry-mass spectrometry reveal key insights into amyloid assembly
Abstract
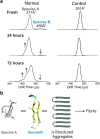
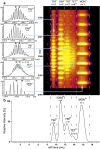
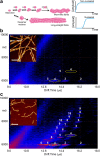
Interfacing ion mobility spectrometry to mass spectrometry (IMS-MS) has enabled mass spectrometric analyses to extend into an extra dimension, providing unrivalled separation and structural characterization of lowly populated species in heterogeneous mixtures. One biological system that has benefitted significantly from such advances is that of amyloid formation. Using IMS-MS, progress has been made into identifying transiently populated monomeric and oligomeric species for a number of different amyloid systems and has led to an enhanced understanding of the mechanism by which small molecules modulate amyloid formation. This review highlights recent advances in this field, which have been accelerated by the commercial availability of IMS-MS instruments. This article is part of a Special Issue entitled: Mass spectrometry in structural biology.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures






References
-
- Dole M., Mack L.L., Hines R.L., Mobley R.C., Ferguson L.D., Alice M.B. Molecular beams of macroions. J. Chem. Phys. 1968;49:2240–2249.
-
- Fenn J.B., Mann M., Meng C.K., Wong S.F., Whitehouse C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. - PubMed
-
- Karas M., Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60:2299–2301. - PubMed
-
- Chowdhury S.K., Katta V., Chait B.T. Probing conformational changes in proteins by mass spectrometry. J. Am. Chem. Soc. 1990;112:9012–9013.
-
- Smith R.D., Light-Wahl K.J. The observation of non-covalent interactions in solution by electrospray ionization mass spectrometry: promise, pitfalls and prognosis. Biol. Mass Spectrom. 1993;22:493–501.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

