Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability
- PMID: 23064114
- PMCID: PMC3597577
- DOI: 10.4161/rna.22360
Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability
Abstract
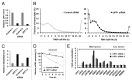
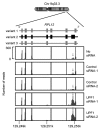
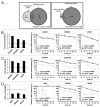
UPF1 eliminates aberrant mRNAs harboring premature termination codons, and regulates the steady-state levels of normal physiological mRNAs. Although genome-wide studies of UPF1 targets performed, previous studies did not distinguish indirect UPF1 targets because they could not determine UPF1-dependent altered RNA stabilities. Here, we measured the decay rates of the whole transcriptome in UPF1-depleted HeLa cells using BRIC-seq, an inhibitor-free method for directly measuring RNA stability. We determined the half-lives and expression levels of 9,229 transcripts. An amount of 785 transcripts were stabilized in UPF1-depleted cells. Among these, the expression levels of 76 transcripts were increased, but those of the other 709 transcripts were not altered. RNA immunoprecipitation showed UPF1 bound to the stabilized transcripts, suggesting that UPF1 directly degrades the 709 transcripts. Many UPF1 targets in this study were newly identified. This study clearly demonstrates that direct determination of RNA stability is a powerful approach for identifying targets of RNA degradation factors.
Keywords: BRIC-seq; RNA decay; RNA-seq; UPF1; nonsense-mediated mRNA decay.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
