SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly
- PMID: 23064147
- PMCID: PMC3501224
- DOI: 10.1038/emboj.2012.280
SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly
Abstract
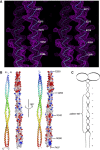
The centriole is a conserved microtubule-based organelle essential for both centrosome formation and cilium biogenesis. Five conserved proteins for centriole duplication have been identified. Two of them, SAS-5 and SAS-6, physically interact with each other and are codependent for their targeting to procentrioles. However, it remains unclear how these two proteins interact at the molecular level. Here, we demonstrate that the short SAS-5 C-terminal domain (residues 390-404) specifically binds to a narrow central region (residues 275-288) of the SAS-6 coiled coil. This was supported by the crystal structure of the SAS-6 coiled-coil domain (CCD), which, together with mutagenesis studies, indicated that the association is mediated by synergistic hydrophobic and electrostatic interactions. The crystal structure also shows a periodic charge pattern along the SAS-6 CCD, which gives rise to an anti-parallel tetramer. Overall, our findings establish the molecular basis of the specific interaction between SAS-5 and SAS-6, and suggest that both proteins individually adopt an oligomeric conformation that is disrupted upon the formation of the hetero-complex to facilitate the correct assembly of the nine-fold symmetric centriole.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 - PubMed
-
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA (2012) Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci 125: 1342–1352 - PubMed
-
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW (2006) Flies without centrioles. Cell 125: 1375–1386 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

