The Armadillo repeat gene ZAK IXIK promotes Arabidopsis early embryo and endosperm development through a distinctive gametophytic maternal effect
- PMID: 23064319
- PMCID: PMC3517234
- DOI: 10.1105/tpc.112.102384
The Armadillo repeat gene ZAK IXIK promotes Arabidopsis early embryo and endosperm development through a distinctive gametophytic maternal effect
Abstract
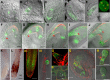
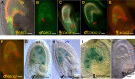
The proper balance of parental genomic contributions to the fertilized embryo and endosperm is essential for their normal growth and development. The characterization of many gametophytic maternal effect (GME) mutants affecting seed development indicates that there are certain classes of genes with a predominant maternal contribution. We present a detailed analysis of the GME mutant zak ixik (zix), which displays delayed and arrested growth at the earliest stages of embryo and endosperm development. ZIX encodes an Armadillo repeat (Arm) protein highly conserved across eukaryotes. Expression studies revealed that ZIX manifests a GME through preferential maternal expression in the early embryo and endosperm. This parent-of-origin-dependent expression is regulated by neither the histone and DNA methylation nor the DNA demethylation pathways known to regulate some other GME mutants. The ZIX protein is localized in the cytoplasm and nucleus of cells in reproductive tissues and actively dividing root zones. The maternal ZIX allele is required for the maternal expression of miniseed3. Collectively, our results reveal a reproductive function of plant Arm proteins in promoting early seed growth, which is achieved through a distinct GME of ZIX that involves mechanisms for maternal allele-specific expression that are independent of the well-established pathways.
Figures
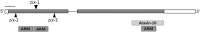





Similar articles
-
SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development.Plant Cell. 2009 Jan;21(1):106-17. doi: 10.1105/tpc.108.064972. Epub 2009 Jan 13. Plant Cell. 2009. PMID: 19141706 Free PMC article.
-
Mutants in the imprinted PICKLE RELATED 2 gene suppress seed abortion of fertilization independent seed class mutants and paternal excess interploidy crosses in Arabidopsis.Plant J. 2017 Apr;90(2):383-395. doi: 10.1111/tpj.13500. Epub 2017 Mar 20. Plant J. 2017. PMID: 28155248
-
Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8.Plant Cell. 2012 Mar;24(3):1000-12. doi: 10.1105/tpc.111.094839. Epub 2012 Mar 16. Plant Cell. 2012. PMID: 22427333 Free PMC article.
-
Epigenetic mechanisms governing seed development in plants.EMBO Rep. 2006 Dec;7(12):1223-7. doi: 10.1038/sj.embor.7400854. EMBO Rep. 2006. PMID: 17139298 Free PMC article. Review.
-
Transcriptional and hormonal signaling control of Arabidopsis seed development.Curr Opin Plant Biol. 2010 Oct;13(5):611-20. doi: 10.1016/j.pbi.2010.08.009. Epub 2010 Sep 25. Curr Opin Plant Biol. 2010. PMID: 20875768 Review.
Cited by
-
Arabidopsis LORELEI, a Maternally Expressed Imprinted Gene, Promotes Early Seed Development.Plant Physiol. 2017 Oct;175(2):758-773. doi: 10.1104/pp.17.00427. Epub 2017 Aug 15. Plant Physiol. 2017. PMID: 28811333 Free PMC article.
-
Armadillo repeat only protein GS10 negatively regulates brassinosteroid signaling to control rice grain size.Plant Physiol. 2023 May 31;192(2):967-981. doi: 10.1093/plphys/kiad117. Plant Physiol. 2023. PMID: 36822628 Free PMC article.
-
Maternal Gametophyte Effects on Seed Development in Maize.Genetics. 2016 Sep;204(1):233-48. doi: 10.1534/genetics.116.191833. Epub 2016 Jul 27. Genetics. 2016. PMID: 27466227 Free PMC article.
-
Spatial and temporal regulation of parent-of-origin allelic expression in the endosperm.Plant Physiol. 2023 Feb 12;191(2):986-1001. doi: 10.1093/plphys/kiac520. Plant Physiol. 2023. PMID: 36437711 Free PMC article.
-
DEAD-BOX RNA HELICASE 27 regulates microRNA biogenesis, zygote division, and stem cell homeostasis.Plant Cell. 2021 Mar 22;33(1):66-84. doi: 10.1093/plcell/koaa001. Plant Cell. 2021. PMID: 33751089 Free PMC article.
References
-
- Amador V., Monte E., García-Martínez J.L., Prat S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106: 343–354 - PubMed
-
- Andreuzza S., Li J., Guitton A.E., Faure J.E., Casanova S., Park J.S., Choi Y., Chen Z., Berger F. (2010). DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development 137: 73–81 - PubMed
-
- Autran D., et al. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 - PubMed
-
- Aw S.J., Hamamura Y., Chen Z., Schnittger A., Berger F. (2010). Sperm entry is sufficient to trigger division of the central cell but the paternal genome is required for endosperm development in Arabidopsis. Development 137: 2683–2690 - PubMed
-
- Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

