Lineage-specific responses of microbial communities to environmental change
- PMID: 23064335
- PMCID: PMC3536104
- DOI: 10.1128/AEM.02226-12
Lineage-specific responses of microbial communities to environmental change
Abstract
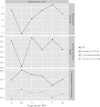
A great challenge facing microbial ecology is how to define ecologically relevant taxonomic units. To address this challenge, we investigated how changing the definition of operational taxonomic units (OTUs) influences the perception of ecological patterns in microbial communities as they respond to a dramatic environmental change. We used pyrosequenced tags of the bacterial V2 16S rRNA region, as well as clone libraries constructed from the cytochrome oxidase C gene ccoN, to provide additional taxonomic resolution for the common freshwater genus Polynucleobacter. At the most highly resolved taxonomic scale, we show that distinct genotypes associated with the abundant Polynucleobacter lineages exhibit divergent spatial patterns and dramatic changes over time, while the also abundant Actinobacteria OTUs are highly coherent. This clearly demonstrates that different bacterial lineages demand different taxonomic definitions to capture ecological patterns. Based on the temporal distribution of highly resolved taxa in the hypolimnion, we demonstrate that change in the population structure of a single genotype can provide additional insight into the mechanisms of community-level responses. These results highlight the importance and feasibility of examining ecological change in microbial communities across taxonomic scales while also providing valuable insight into the ecological characteristics of ecologically coherent groups in this system.
Figures



References
-
- Curtis TP, Sloan WT. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221–226 - PubMed
-
- Dobrindt U, Hacker J. 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4:550–557 - PubMed
-
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, Baurain D. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9:e1000602 doi:10.1371/journal.pbio.1000602 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

