Genotype calling and haplotyping in parent-offspring trios
- PMID: 23064751
- PMCID: PMC3530674
- DOI: 10.1101/gr.142455.112
Genotype calling and haplotyping in parent-offspring trios
Abstract
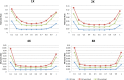
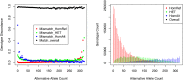
Emerging sequencing technologies allow common and rare variants to be systematically assayed across the human genome in many individuals. In order to improve variant detection and genotype calling, raw sequence data are typically examined across many individuals. Here, we describe a method for genotype calling in settings where sequence data are available for unrelated individuals and parent-offspring trios and show that modeling trio information can greatly increase the accuracy of inferred genotypes and haplotypes, especially on low to modest depth sequencing data. Our method considers both linkage disequilibrium (LD) patterns and the constraints imposed by family structure when assigning individual genotypes and haplotypes. Using simulations, we show that trios provide higher genotype calling accuracy across the frequency spectrum, both overall and at hard-to-call heterozygous sites. In addition, trios provide greatly improved phasing accuracy--improving the accuracy of downstream analyses (such as genotype imputation) that rely on phased haplotypes. To further evaluate our approach, we analyzed data on the first 508 individuals sequenced by the SardiNIA sequencing project. Our results show that our method reduces the genotyping error rate by 50% compared with analysis using existing methods that ignore family structure. We anticipate our method will facilitate genotype calling and haplotype inference for many ongoing sequencing projects.
Figures

References
-
- Cirulli ET, Goldstein DB 2010. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 11: 415–425 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
