Breakpoint-specific multiplex polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia
- PMID: 23065506
- PMCID: PMC3659991
- DOI: 10.3324/haematol.2012.073965
Breakpoint-specific multiplex polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia
Abstract
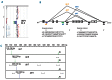
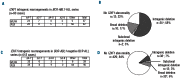
Deletion of the Ikaros (IKZF1) gene is an oncogenic lesion frequently associated with BCR-ABL1-positive acute lymphoblastic leukemias. It is also found in a fraction of BCR-ABL1-negative B-cell precursor acute lymphoblastic leukemias, and early studies showed it was associated with a higher risk of relapse. Therefore, screening tools are needed for evaluation in treatment protocols and possible inclusion in risk stratification. Besides monosomy 7 and large 7p abnormalities encompassing IKZF1, most IKZF1 alterations are short, intragenic deletions. Based on cohorts of patients, we mapped the microdeletion breakpoints and developed a breakpoint-specific fluorescent multiplex polymerase chain reaction that allows detection of recurrent intragenic deletions. This sensitive test could also detect IKZF1 subclonal deletions, whose prognostic significance should be evaluated. Moreover, we show that consensus breakpoint sequences can be used as clonal markers to monitor minimal residual disease. This paper could be useful for translational studies and in clinical management of BCP-ALL.
Figures

References
-
- Moorman AV. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. 2012;26(3):123-35 - PubMed
-
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758-64 - PubMed
-
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110-4 - PubMed
-
- Waanders E, van der Velden VH, van der Schoot CE, van Leeuwen FN, van Reijmersdal SV, de Haas V, et al. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia. 2011;25(2):254-8 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

