PlantRNA, a database for tRNAs of photosynthetic eukaryotes
- PMID: 23066098
- PMCID: PMC3531208
- DOI: 10.1093/nar/gks935
PlantRNA, a database for tRNAs of photosynthetic eukaryotes
Abstract
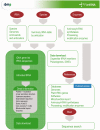
PlantRNA database (http://plantrna.ibmp.cnrs.fr/) compiles transfer RNA (tRNA) gene sequences retrieved from fully annotated plant nuclear, plastidial and mitochondrial genomes. The set of annotated tRNA gene sequences has been manually curated for maximum quality and confidence. The novelty of this database resides in the inclusion of biological information relevant to the function of all the tRNAs entered in the library. This includes 5'- and 3'-flanking sequences, A and B box sequences, region of transcription initiation and poly(T) transcription termination stretches, tRNA intron sequences, aminoacyl-tRNA synthetases and enzymes responsible for tRNA maturation and modification. Finally, data on mitochondrial import of nuclear-encoded tRNAs as well as the bibliome for the respective tRNAs and tRNA-binding proteins are also included. The current annotation concerns complete genomes from 11 organisms: five flowering plants (Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Medicago truncatula and Brachypodium distachyon), a moss (Physcomitrella patens), two green algae (Chlamydomonas reinhardtii and Ostreococcus tauri), one glaucophyte (Cyanophora paradoxa), one brown alga (Ectocarpus siliculosus) and a pennate diatom (Phaeodactylum tricornutum). The database will be regularly updated and implemented with new plant genome annotations so as to provide extensive information on tRNA biology to the research community.
Figures

Similar articles
-
PlantRNA 2.0: an updated database dedicated to tRNAs of photosynthetic eukaryotes.Plant J. 2022 Nov;112(4):1112-1119. doi: 10.1111/tpj.15997. Epub 2022 Oct 17. Plant J. 2022. PMID: 36196656
-
PLMItRNA, a database on the heterogeneous genetic origin of mitochondrial tRNA genes and tRNAs in photosynthetic eukaryotes.Nucleic Acids Res. 2003 Jan 1;31(1):436-8. doi: 10.1093/nar/gkg080. Nucleic Acids Res. 2003. PMID: 12520044 Free PMC article.
-
Extensive import of nucleus-encoded tRNAs into chloroplasts of the photosynthetic lycophyte, Selaginella kraussiana.Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2412221121. doi: 10.1073/pnas.2412221121. Epub 2024 Nov 6. Proc Natl Acad Sci U S A. 2024. PMID: 39503889 Free PMC article.
-
Mitochondrial tRNA import and its consequences for mitochondrial translation.Annu Rev Biochem. 2011;80:1033-53. doi: 10.1146/annurev-biochem-060109-092838. Annu Rev Biochem. 2011. PMID: 21417719 Review.
-
DNA Import into Mitochondria.Biochemistry (Mosc). 2016 Oct;81(10):1044-1056. doi: 10.1134/S0006297916100035. Biochemistry (Mosc). 2016. PMID: 27908230 Review.
Cited by
-
tRNA derived small RNAs-Small players with big roles.Front Genet. 2022 Sep 19;13:997780. doi: 10.3389/fgene.2022.997780. eCollection 2022. Front Genet. 2022. PMID: 36199575 Free PMC article. Review.
-
Arabidopsis tRNA-derived fragments as potential modulators of translation.RNA Biol. 2020 Aug;17(8):1137-1148. doi: 10.1080/15476286.2020.1722514. Epub 2020 Feb 5. RNA Biol. 2020. PMID: 31994438 Free PMC article.
-
Characterization of Frond and Flower Development and Identification of FT and FD Genes From Duckweed Lemna aequinoctialis Nd.Front Plant Sci. 2021 Oct 11;12:697206. doi: 10.3389/fpls.2021.697206. eCollection 2021. Front Plant Sci. 2021. PMID: 34707626 Free PMC article.
-
Protein-only RNase P function in Escherichia coli: viability, processing defects and differences between PRORP isoenzymes.Nucleic Acids Res. 2017 Jul 7;45(12):7441-7454. doi: 10.1093/nar/gkx405. Nucleic Acids Res. 2017. PMID: 28499021 Free PMC article.
-
Extensive profiling of the expressions of tRNAs and tRNA-derived fragments (tRFs) reveals the complexities of tRNA and tRF populations in plants.Sci China Life Sci. 2021 Apr;64(4):495-511. doi: 10.1007/s11427-020-1891-8. Epub 2021 Feb 8. Sci China Life Sci. 2021. PMID: 33569675
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

