LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets
- PMID: 23066108
- PMCID: PMC3526318
- DOI: 10.1093/nar/gks918
LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets
Abstract
The study of cell-population heterogeneity in a range of biological systems, from viruses to bacterial isolates to tumor samples, has been transformed by recent advances in sequencing throughput. While the high-coverage afforded can be used, in principle, to identify very rare variants in a population, existing ad hoc approaches frequently fail to distinguish true variants from sequencing errors. We report a method (LoFreq) that models sequencing run-specific error rates to accurately call variants occurring in <0.05% of a population. Using simulated and real datasets (viral, bacterial and human), we show that LoFreq has near-perfect specificity, with significantly improved sensitivity compared with existing methods and can efficiently analyze deep Illumina sequencing datasets without resorting to approximations or heuristics. We also present experimental validation for LoFreq on two different platforms (Fluidigm and Sequenom) and its application to call rare somatic variants from exome sequencing datasets for gastric cancer. Source code and executables for LoFreq are freely available at http://sourceforge.net/projects/lofreq/.
Figures
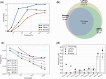
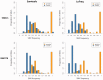

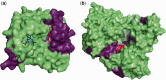
References
-
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Die Naturwissenschaften. 1971;58:465–523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

