Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression?
- PMID: 23069990
- PMCID: PMC3708292
- DOI: 10.1007/s00018-012-1186-z
Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression?
Abstract
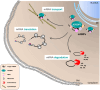
The insulin-like growth factor-2 mRNA-binding proteins 1, 2, and 3 (IGF2BP1, IGF2BP2, IGF2BP3) belong to a conserved family of RNA-binding, oncofetal proteins. Several studies have shown that these proteins act in various important aspects of cell function, such as cell polarization, migration, morphology, metabolism, proliferation and differentiation. In this review, we discuss the IGF2BP family's role in cancer biology and how this correlates with their proposed functions during embryogenesis. IGF2BPs are mainly expressed in the embryo, in contrast with comparatively lower or negotiable levels in adult tissues. IGF2BP1 and IGF2BP3 have been found to be re-expressed in several aggressive cancer types. Control of IGF2BPs' expression is not well understood; however, let-7 microRNAs, β-catenin (CTNNB1) and MYC have been proposed to be involved in their regulation. In contrast to many other RNA-binding proteins, IGF2BPs are almost exclusively observed in the cytoplasm where they associate with target mRNAs in cytoplasmic ribonucleoprotein complexes (mRNPs). During development, IGF2BPs are required for proper nerve cell migration and morphological development, presumably involving the control of cytoskeletal remodeling and dynamics, respectively. Likewise, IGF2BPs modulate cell polarization, adhesion and migration in tumor-derived cells. Moreover, they are highly associated with cancer metastasis and the expression of oncogenic factors (KRAS, MYC and MDR1). However, a pro-metastatic role of IGF2BPs remains controversial due to the lack of 'classical' in vivo studies. Nonetheless, IGF2BPs could provide valuable targets in cancer treatment with many of their in vivo roles to be fully elucidated.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

