The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme
- PMID: 23071111
- PMCID: PMC3504732
- DOI: 10.1074/jbc.M112.421776
The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme
Abstract
Background: Phosphatidate phosphatase (PAP) plays diverse roles in lipid metabolism and cell signaling.
Results: A novel yeast PAP is identified as the actin patch protein encoded by APP1.
Conclusion: APP1 and other known genes (PAH1, DPP1, LPP1) are responsible for all detectable PAP activity in yeast.
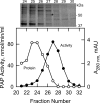
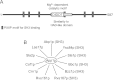
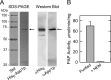
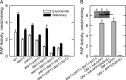
Significance: Identification of App1p as a PAP enzyme will facilitate the understanding of its cellular function. Phosphatidate phosphatase (PAP) catalyzes the dephosphorylation of phosphatidate to yield diacylglycerol. In the yeast Saccharomyces cerevisiae, PAP is encoded by PAH1, DPP1, and LPP1. The presence of PAP activity in the pah1Δ dpp1Δ lpp1Δ triple mutant indicated another gene(s) encoding the enzyme. We purified PAP from the pah1Δ dpp1Δ lpp1Δ triple mutant by salt extraction of mitochondria followed by chromatography with DE52, Affi-Gel Blue, phenyl-Sepharose, MonoQ, and Superdex 200. Liquid chromatography/tandem mass spectrometry analysis of a PAP-enriched sample revealed multiple putative phosphatases. By analysis of PAP activity in mutants lacking each of the proteins, we found that APP1, a gene whose molecular function has been unknown, confers ~30% PAP activity of wild type cells. The overexpression of APP1 in the pah1Δ dpp1Δ lpp1Δ mutant exhibited a 10-fold increase in PAP activity. The PAP activity shown by App1p heterologously expressed in Escherichia coli confirmed that APP1 is the structural gene for the enzyme. Introduction of the app1Δ mutation into the pah1Δ dpp1Δ lpp1Δ triple mutant resulted in a complete loss of PAP activity, indicating that distinct PAP enzymes in S. cerevisiae are encoded by APP1, PAH1, DPP1, and LPP1. Lipid analysis of cells lacking the PAP genes, singly or in combination, showed that Pah1p is the only PAP involved in the synthesis of triacylglycerol as well as in the regulation of phospholipid synthesis. App1p, which shows interactions with endocytic proteins, may play a role in vesicular trafficking through its PAP activity.
Figures

Similar articles
-
PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae.J Biol Chem. 2013 Dec 13;288(50):35781-92. doi: 10.1074/jbc.M113.525766. Epub 2013 Nov 6. J Biol Chem. 2013. PMID: 24196957 Free PMC article.
-
Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase.J Biol Chem. 1998 Jun 5;273(23):14331-8. doi: 10.1074/jbc.273.23.14331. J Biol Chem. 1998. PMID: 9603941
-
Discoveries of the phosphatidate phosphatase genes in yeast published in the Journal of Biological Chemistry.J Biol Chem. 2019 Feb 1;294(5):1681-1689. doi: 10.1074/jbc.TM118.004159. Epub 2018 Jul 30. J Biol Chem. 2019. PMID: 30061152 Free PMC article. Review.
-
Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis.J Biol Chem. 2017 Aug 11;292(32):13230-13242. doi: 10.1074/jbc.M117.801720. Epub 2017 Jul 3. J Biol Chem. 2017. PMID: 28673963 Free PMC article.
-
Phosphatidate phosphatase, a key regulator of lipid homeostasis.Biochim Biophys Acta. 2013 Mar;1831(3):514-22. doi: 10.1016/j.bbalip.2012.08.006. Epub 2012 Aug 14. Biochim Biophys Acta. 2013. PMID: 22910056 Free PMC article. Review.
Cited by
-
A genome-wide CRISPR screen identifies GRA38 as a key regulator of lipid homeostasis during Toxoplasma gondii adaptation to lipid-rich conditions.Res Sq [Preprint]. 2025 Apr 18:rs.3.rs-6436164. doi: 10.21203/rs.3.rs-6436164/v1. Res Sq. 2025. PMID: 40321756 Free PMC article. Preprint.
-
Involvement of phosphatidate phosphatase in the biosynthesis of triacylglycerols in Chlamydomonas reinhardtii.J Zhejiang Univ Sci B. 2013 Dec;14(12):1121-31. doi: 10.1631/jzus.B1300180. J Zhejiang Univ Sci B. 2013. PMID: 24302712 Free PMC article.
-
Engineering yeast for tailored fatty acid profiles.Appl Microbiol Biotechnol. 2025 Apr 22;109(1):101. doi: 10.1007/s00253-025-13487-1. Appl Microbiol Biotechnol. 2025. PMID: 40263140 Free PMC article. Review.
-
The Inner Nuclear Membrane Is a Metabolically Active Territory that Generates Nuclear Lipid Droplets.Cell. 2018 Jul 26;174(3):700-715.e18. doi: 10.1016/j.cell.2018.05.047. Epub 2018 Jun 21. Cell. 2018. PMID: 29937227 Free PMC article.
-
PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae.J Biol Chem. 2013 Dec 13;288(50):35781-92. doi: 10.1074/jbc.M113.525766. Epub 2013 Nov 6. J Biol Chem. 2013. PMID: 24196957 Free PMC article.
References
-
- Smith S. W., Weiss S. B., Kennedy E. P. (1957) The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 - PubMed
-
- Kennedy E. P., Weiss S. B. (1956) The function of cytidine coenzyme in the biosynthesis of phospholipids. J. Biol. Chem. 222, 193–214 - PubMed
-
- Borkenhagen L. F., Kennedy E. P. (1957) The enzymatic synthesis of cytidine diphosphate choline. J. Biol. Chem. 227, 951–962 - PubMed
-
- Weiss S. B., Smith S. W., Kennedy E. P. (1958) The enzymatic formation of lecithin from cytidine diphosphate choline and d-1,2-diglyceride. J. Biol. Chem. 231, 53–64 - PubMed
-
- Kennedy E. P. (1956) The synthesis of cytidine diphosphate choline, cytidine diphosphate ethanolamine, and related compounds. J. Biol. Chem. 222, 185–191 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

