IGHV-unmutated and IGHV-mutated chronic lymphocytic leukemia cells produce activation-induced deaminase protein with a full range of biologic functions
- PMID: 23071276
- PMCID: PMC3520620
- DOI: 10.1182/blood-2012-08-449744
IGHV-unmutated and IGHV-mutated chronic lymphocytic leukemia cells produce activation-induced deaminase protein with a full range of biologic functions
Abstract
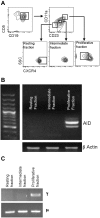
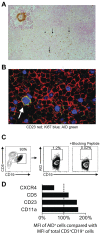
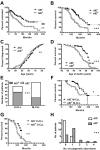
Clonal evolution occurs during the course of chronic lymphocytic leukemia (CLL) and activation-induced deaminase (AID) could influence this process. However, this possibility has been questioned in CLL because the number of circulating AID mRNA(+) cells is exceedingly low; synthesis of AID protein by blood CLL cells has not been demonstrated; the full range of AID functions is lacking in unmutated CLL (U-CLL), and no prospective analysis linking AID expression and disease severity has been reported. The results of the present study show that circulating CLL cells and those within secondary lymphoid tissues can make AID mRNA and protein. This production is related to cell division because more AID mRNA was detected in recently divided cells and AID protein was limited to the dividing fraction and was up-regulated on induction of cell division. AID protein was functional because AID(+) dividing cells exhibited more double-stranded DNA breaks, IGH class switching, and new IGHV-D-J mutations. Each of these actions was documented in U-CLL and mutated CLL (M-CLL). Furthermore, AID protein was associated with worse patient outcome and adverse cytogenetics. We conclude that the production of fully functional AID protein by U-CLL and M-CLL cells could be involved in clonal evolution of the disease.
Figures



References
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. - PubMed
-
- Shanafelt TD, Witzig TE, Fink SR, et al. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24(19):4634–4641. - PubMed
-
- Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

