Provirophages and transpovirons as the diverse mobilome of giant viruses
- PMID: 23071316
- PMCID: PMC3497776
- DOI: 10.1073/pnas.1208835109
Provirophages and transpovirons as the diverse mobilome of giant viruses
Abstract
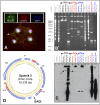
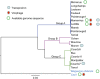
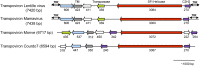
A distinct class of infectious agents, the virophages that infect giant viruses of the Mimiviridae family, has been recently described. Here we report the simultaneous discovery of a giant virus of Acanthamoeba polyphaga (Lentille virus) that contains an integrated genome of a virophage (Sputnik 2), and a member of a previously unknown class of mobile genetic elements, the transpovirons. The transpovirons are linear DNA elements of ~7 kb that encompass six to eight protein-coding genes, two of which are homologous to virophage genes. Fluorescence in situ hybridization showed that the free form of the transpoviron replicates within the giant virus factory and accumulates in high copy numbers inside giant virus particles, Sputnik 2 particles, and amoeba cytoplasm. Analysis of deep-sequencing data showed that the virophage and the transpoviron can integrate in nearly any place in the chromosome of the giant virus host and that, although less frequently, the transpoviron can also be linked to the virophage chromosome. In addition, integrated fragments of transpoviron DNA were detected in several giant virus and Sputnik genomes. Analysis of 19 Mimivirus strains revealed three distinct transpovirons associated with three subgroups of Mimiviruses. The virophage, the transpoviron, and the previously identified self-splicing introns and inteins constitute the complex, interconnected mobilome of the giant viruses and are likely to substantially contribute to interviral gene transfer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Virology: giant viruses--movers and shakers.Nat Rev Microbiol. 2012 Dec;10(12):803. doi: 10.1038/nrmicro2927. Nat Rev Microbiol. 2012. PMID: 23154257 No abstract available.
References
-
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: The agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–732. - PubMed
-
- Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303(5664):1626–1632. - PubMed
-
- Siefert JL. Defining the mobilome. Methods Mol Biol. 2009;532:13–27. - PubMed
-
- Sun AJ, et al. Functional evaluation of the role of reticuloendotheliosis virus long terminal repeat (LTR) integrated into the genome of a field strain of Marek’s disease virus. Virology. 2010;397(2):270–276. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

