SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas
- PMID: 23071531
- PMCID: PMC3468603
- DOI: 10.1371/journal.pone.0045950
SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas
Erratum in
- PLoS One. 2013;8(5). doi:10.1371/annotation/27807b78-9c79-414a-a47e-fb3eca621be4. Verelle, Pierre [corrected to Verrelle, Pierre]
Abstract
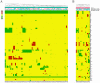
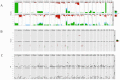
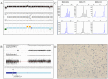
Anaplastic oligodendrogliomas (AOD) are rare glial tumors in adults with relative homogeneous clinical, radiological and histological features at the time of diagnosis but dramatically various clinical courses. Studies have identified several molecular abnormalities with clinical or biological relevance to AOD (e.g. t(1;19)(q10;p10), IDH1, IDH2, CIC and FUBP1 mutations).To better characterize the clinical and biological behavior of this tumor type, the creation of a national multicentric network, named "Prise en charge des OLigodendrogliomes Anaplasiques (POLA)," has been supported by the Institut National du Cancer (InCA). Newly diagnosed and centrally validated AOD patients and their related biological material (tumor and blood samples) were prospectively included in the POLA clinical database and tissue bank, respectively.At the molecular level, we have conducted a high-resolution single nucleotide polymorphism array analysis, which included 83 patients. Despite a careful central pathological review, AOD have been found to exhibit heterogeneous genomic features. A total of 82% of the tumors exhibited a 1p/19q-co-deletion, while 18% harbor a distinct chromosome pattern. Novel focal abnormalities, including homozygously deleted, amplified and disrupted regions, have been identified. Recurring copy neutral losses of heterozygosity (CNLOH) inducing the modulation of gene expression have also been discovered. CNLOH in the CDKN2A locus was associated with protein silencing in 1/3 of the cases. In addition, FUBP1 homozygous deletion was detected in one case suggesting a putative tumor suppressor role of FUBP1 in AOD.Our study showed that the genomic and pathological analyses of AOD are synergistic in detecting relevant clinical and biological subgroups of AOD.
Conflict of interest statement
Figures

Similar articles
-
CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas.Acta Neuropathol. 2012 Jun;123(6):853-60. doi: 10.1007/s00401-012-0993-5. Epub 2012 May 17. Acta Neuropathol. 2012. PMID: 22588899
-
Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene.Clin Cancer Res. 2005 Feb 1;11(3):1119-28. Clin Cancer Res. 2005. PMID: 15709179
-
Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: a clinicopathological study using fluorescence in situ hybridization.Neuropathology. 2007 Feb;27(1):10-20. doi: 10.1111/j.1440-1789.2006.00735.x. Neuropathology. 2007. PMID: 17319279
-
Oligodendrogliomas: new insights from the genetics and perspectives.Curr Opin Oncol. 2012 Nov;24(6):687-93. doi: 10.1097/CCO.0b013e328357f4ea. Curr Opin Oncol. 2012. PMID: 22913971 Review.
-
Molecular background of oligodendroglioma: 1p/19q, IDH, TERT, CIC and FUBP1.CNS Oncol. 2015;4(5):287-94. doi: 10.2217/cns.15.32. Epub 2015 Nov 6. CNS Oncol. 2015. PMID: 26545048 Free PMC article. Review.
Cited by
-
Machine Learning for Better Prognostic Stratification and Driver Gene Identification Using Somatic Copy Number Variations in Anaplastic Oligodendroglioma.Oncologist. 2018 Dec;23(12):1500-1510. doi: 10.1634/theoncologist.2017-0495. Epub 2018 Jul 17. Oncologist. 2018. PMID: 30018130 Free PMC article.
-
Molecular pathogenesis of B-cell posttransplant lymphoproliferative disorder: what do we know so far?Clin Dev Immunol. 2013;2013:150835. doi: 10.1155/2013/150835. Epub 2013 Apr 14. Clin Dev Immunol. 2013. PMID: 23690819 Free PMC article. Review.
-
Deep sequencing of a recurrent oligodendroglioma and the derived xenografts reveals new insights into the evolution of human oligodendroglioma and candidate driver genes.Oncotarget. 2019 Jun 4;10(38):3641-3653. doi: 10.18632/oncotarget.26950. eCollection 2019 Jun 4. Oncotarget. 2019. PMID: 31217899 Free PMC article.
-
Allelic loss of 9p21.3 is a prognostic factor in 1p/19q codeleted anaplastic gliomas.Neurology. 2015 Oct 13;85(15):1325-31. doi: 10.1212/WNL.0000000000002014. Epub 2015 Sep 18. Neurology. 2015. PMID: 26385879 Free PMC article.
-
The prognostic impact of CDKN2A/B hemizygous deletions in IDH-mutant glioma.Neuro Oncol. 2025 Mar 7;27(3):743-754. doi: 10.1093/neuonc/noae238. Neuro Oncol. 2025. PMID: 39530475
References
-
- Central Brain Tumor Registry of the United States (n.d.). Available:http://www.cbtrus.org/. Accessed 2012 Jun 10.
-
- Rigau V, Zouaoui S, Mathieu-Daudé H, Darlix A, Maran A, et al. (2011) French brain tumor database: 5-year histological results on 25 756 cases. Brain Pathol 21: 633–644 doi:10.1111/j.1750-3639.2011.00491.x - DOI - PMC - PubMed
-
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, et al. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97–109 doi:10.1007/s00401-007-0243-4 - DOI - PMC - PubMed
-
- Cairncross G, Berkey B, Shaw E, Jenkins R, Scheithauer B, et al. (2006) Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol 24: 2707–2714 doi:10.1200/JCO.2005.04.3414 - DOI - PubMed
-
- van den Bent MJ, Carpentier AF, Brandes AA, Sanson M, Taphoorn MJB, et al. (2006) Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol 24: 2715–2722 doi:10.1200/JCO.2005.04.6078 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

