Gene-gene interaction and functional impact of polymorphisms on innate immune genes in controlling Plasmodium falciparum blood infection level
- PMID: 23071570
- PMCID: PMC3470565
- DOI: 10.1371/journal.pone.0046441
Gene-gene interaction and functional impact of polymorphisms on innate immune genes in controlling Plasmodium falciparum blood infection level
Abstract
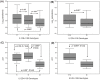
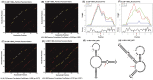
Genetic variations in toll-like receptors and cytokine genes of the innate immune pathways have been implicated in controlling parasite growth and the pathogenesis of Plasmodium falciparum mediated malaria. We previously published genetic association of TLR4 non-synonymous and TNF-α promoter polymorphisms with P.falciparum blood infection level and here we extend the study considerably by (i) investigating genetic dependence of parasite-load on interleukin-12B polymorphisms, (ii) reconstructing gene-gene interactions among candidate TLRs and cytokine loci, (iii) exploring genetic and functional impact of epistatic models and (iv) providing mechanistic insights into functionality of disease-associated regulatory polymorphisms. Our data revealed that carriage of AA (P = 0.0001) and AC (P = 0.01) genotypes of IL12B 3'UTR polymorphism was associated with a significant increase of mean log-parasitemia relative to rare homozygous genotype CC. Presence of IL12B+1188 polymorphism in five of six multifactor models reinforced its strong genetic impact on malaria phenotype. Elevation of genetic risk in two-component models compared to the corresponding single locus and reduction of IL12B (2.2 fold) and lymphotoxin-α (1.7 fold) expressions in patients'peripheral-blood-mononuclear-cells under TLR4Thr399Ile risk genotype background substantiated the role of Multifactor Dimensionality Reduction derived models. Marked reduction of promoter activity of TNF-α risk haplotype (C-C-G-G) compared to wild-type haplotype (T-C-G-G) with (84%) and without (78%) LPS stimulation and the loss of binding of transcription factors detected in-silico supported a causal role of TNF-1031. Significantly lower expression of IL12B+1188 AA (5 fold) and AC (9 fold) genotypes compared to CC and under-representation (P = 0.0048) of allele A in transcripts of patients' PBMCs suggested an Allele-Expression-Imbalance. Allele (A+1188C) dependent differential stability (2 fold) of IL12B-transcripts upon actinomycin-D treatment and observed structural modulation (P = 0.013) of RNA-ensemble were the plausible explanations for AEI. In conclusion, our data provides functional support to the hypothesis that de-regulated receptor-cytokine axis of innate immune pathway influences blood infection level in P. falciparum malaria.
Conflict of interest statement
Figures




Similar articles
-
Genetic association of Toll-like-receptor 4 and tumor necrosis factor-alpha polymorphisms with Plasmodium falciparum blood infection levels.Infect Genet Evol. 2010 Jul;10(5):686-96. doi: 10.1016/j.meegid.2010.03.008. Epub 2010 Mar 20. Infect Genet Evol. 2010. PMID: 20307689
-
Haplotypes of IL12B promoter polymorphisms condition susceptibility to severe malaria and functional changes in cytokine levels in Thai adults.Immunogenetics. 2010 Jun;62(6):345-56. doi: 10.1007/s00251-010-0439-y. Epub 2010 Apr 13. Immunogenetics. 2010. PMID: 20387064 Free PMC article.
-
IL12B polymorphisms are linked but not associated with Plasmodium falciparum parasitemia: a familial study in Burkina Faso.Genes Immun. 2008 Jul;9(5):405-11. doi: 10.1038/gene.2008.31. Epub 2008 May 1. Genes Immun. 2008. PMID: 18449199
-
A meta-analysis of TLR4 and TLR9 SNPs implicated in severe malaria.Rev Soc Bras Med Trop. 2017 Mar-Apr;50(2):153-160. doi: 10.1590/0037-8682-0475-2016. Rev Soc Bras Med Trop. 2017. PMID: 28562749 Review.
-
Malaria blood-stage infection and its control by the immune system.Folia Biol (Praha). 2000;46(6):210-8. Folia Biol (Praha). 2000. PMID: 11140853 Review.
Cited by
-
Two susceptibility loci to Takayasu arteritis reveal a synergistic role of the IL12B and HLA-B regions in a Japanese population.Am J Hum Genet. 2013 Aug 8;93(2):289-97. doi: 10.1016/j.ajhg.2013.05.024. Epub 2013 Jul 3. Am J Hum Genet. 2013. PMID: 23830516 Free PMC article.
-
Cord Blood IL-12 Confers Protection to Clinical Malaria in Early Childhood Life.Sci Rep. 2018 Jul 18;8(1):10860. doi: 10.1038/s41598-018-29179-y. Sci Rep. 2018. PMID: 30022038 Free PMC article.
-
A roadmap to multifactor dimensionality reduction methods.Brief Bioinform. 2016 Mar;17(2):293-308. doi: 10.1093/bib/bbv038. Epub 2015 Jun 24. Brief Bioinform. 2016. PMID: 26108231 Free PMC article. Review.
-
Diversity analysis of MSP1 identifies conserved epitope organization in block 2 amidst high sequence variability in Indian Plasmodium falciparum isolates.Malar J. 2018 Dec 3;17(1):447. doi: 10.1186/s12936-018-2592-y. Malar J. 2018. PMID: 30509263 Free PMC article.
-
Variants in two gene members of the TNF ligand superfamily and hepatitis C virus chronic disease.Gastroenterol Hepatol Bed Bench. 2018 Winter;11(Suppl 1):S66-S72. Gastroenterol Hepatol Bed Bench. 2018. PMID: 30774809 Free PMC article.
References
-
- World Health Organization (2011) World Malaria Report. GenevaSwitzerland: World Health Organization.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

