Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato
- PMID: 23071630
- PMCID: PMC3465262
- DOI: 10.1371/journal.pone.0046763
Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato
Abstract
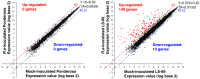
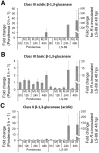
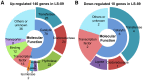
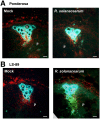
Bacterial wilt, caused by the soil-borne bacterium Ralstonia solanacearum, is a lethal disease of tomato, but the molecular mechanisms of the host resistance responses to R. solanacearum remain unclear. In this study, we report the first work describing the transcriptome of cultivar resistance and susceptible tomato cultivar after inoculation with R. solanacearum. To elucidate the characteristics of resistance early in the interaction, we analyzed microarrays for resistant cultivar LS-89 and susceptible cultivar Ponderosa 1 day after stem inoculation. No change in gene expression was detected for Ponderosa, but expression levels of over 140 genes, including pathogenesis-related, hormone signaling and lignin biosynthesis genes, increased in LS-89. Expression of β-1,3-glucanase genes increased substantially. In an immunohistochemical study, glucanase in LS-89 accumulated in the xylem and pith tissues surrounding xylem vessels filled with R. solanacearum. The expression of these genes also increased in four other resistant cultivars, but changed little in four susceptible cultivars in response to R. solanacearum, suggesting that similar reactions occur in other cultivars. These gene expression profiles will serve as fundamental information to elucidate the molecular mechanisms in the resistance response to R. solanacearum in tomato.
Conflict of interest statement
Figures



Similar articles
-
Resistance against Ralstonia solanacearum in tomato depends on the methionine cycle and the γ-aminobutyric acid metabolic pathway.Plant J. 2019 Mar;97(6):1032-1047. doi: 10.1111/tpj.14175. Epub 2019 Jan 5. Plant J. 2019. PMID: 30480846
-
Pleiotropic Phenotypes of the Tomato diageotropica Mutant Enable Resistance to Ralstonia solanacearum.Mol Plant Microbe Interact. 2025 Jul;38(4):566-578. doi: 10.1094/MPMI-10-24-0123-R. Epub 2025 Aug 11. Mol Plant Microbe Interact. 2025. PMID: 40281390
-
A Single Regulator Mediates Strategic Switching between Attachment/Spread and Growth/Virulence in the Plant Pathogen Ralstonia solanacearum.mBio. 2017 Sep 26;8(5):e00895-17. doi: 10.1128/mBio.00895-17. mBio. 2017. PMID: 28951474 Free PMC article.
-
Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era.Mol Plant Pathol. 2013 Sep;14(7):651-62. doi: 10.1111/mpp.12038. Epub 2013 May 30. Mol Plant Pathol. 2013. PMID: 23718203 Free PMC article. Review.
-
Strategies utilized by plants to defend against Ralstonia solanacearum.Front Plant Sci. 2025 May 26;16:1510177. doi: 10.3389/fpls.2025.1510177. eCollection 2025. Front Plant Sci. 2025. PMID: 40491817 Free PMC article. Review.
Cited by
-
The Bacterial Wilt Reservoir Host Solanum dulcamara Shows Resistance to Ralstonia solanacearum Infection.Front Plant Sci. 2021 Nov 10;12:755708. doi: 10.3389/fpls.2021.755708. eCollection 2021. Front Plant Sci. 2021. PMID: 34868145 Free PMC article.
-
Lignin: characterization of a multifaceted crop component.ScientificWorldJournal. 2013 Nov 14;2013:436517. doi: 10.1155/2013/436517. ScientificWorldJournal. 2013. PMID: 24348159 Free PMC article. Review.
-
Tapping the rhizosphere metabolites for the prebiotic control of soil-borne bacterial wilt disease.Nat Commun. 2023 Jul 26;14(1):4497. doi: 10.1038/s41467-023-40184-2. Nat Commun. 2023. PMID: 37495619 Free PMC article.
-
Comparison of the transcriptomes of ginger (Zingiber officinale Rosc.) and mango ginger (Curcuma amada Roxb.) in response to the bacterial wilt infection.PLoS One. 2014 Jun 18;9(6):e99731. doi: 10.1371/journal.pone.0099731. eCollection 2014. PLoS One. 2014. PMID: 24940878 Free PMC article.
-
Induced defense strategies of plants against Ralstonia solanacearum.Front Microbiol. 2023 Jan 26;14:1059799. doi: 10.3389/fmicb.2023.1059799. eCollection 2023. Front Microbiol. 2023. PMID: 36778883 Free PMC article. Review.
References
-
- Hayward AC (1991) Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum . Annu Rev Phytopathol 29: 65–87. - PubMed
-
- Lee JM, Bang HJ, Ham HS (1998) Grafting of vegetables. J Japan Soc Hort Sci 67: 1098–1104.
-
- Hanson PM, Licardo O, Hanudin, Wang JF, Chen JT (1998) Diallel analysis of bacterial wilt resistance in tomato derived from different sources. Plant Dis 82: 74–78. - PubMed
-
- Lebeau A, Daunay MC, Frary A, Palloix A, Wang JF, et al. (2011) Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 101: 154–165. - PubMed
-
- Grimault V, Prior P (1993) Bacterial wilt resistance in tomato associated with tolerance of vascular tissues to Pseudomonas solanacearum . Plant Pathol 42: 589–594.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

