Upper airways microbiota in antibiotic-naïve wheezing and healthy infants from the tropics of rural Ecuador
- PMID: 23071640
- PMCID: PMC3465279
- DOI: 10.1371/journal.pone.0046803
Upper airways microbiota in antibiotic-naïve wheezing and healthy infants from the tropics of rural Ecuador
Abstract
Background: Observations that the airway microbiome is disturbed in asthma may be confounded by the widespread use of antibiotics and inhaled steroids. We have therefore examined the oropharyngeal microbiome in early onset wheezing infants from a rural area of tropical Ecuador where antibiotic usage is minimal and glucocorticoid usage is absent.
Materials and methods: We performed pyrosequencing of amplicons of the polymorphic bacterial 16S rRNA gene from oropharyngeal samples from 24 infants with non-infectious early onset wheezing and 24 healthy controls (average age 10.2 months). We analyzed microbial community structure and differences between cases and controls by QIIME software.
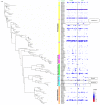
Results: We obtained 76,627 high quality sequences classified into 182 operational taxonomic units (OTUs). Firmicutes was the most common and diverse phylum (71.22% of sequences) with Streptococcus being the most common genus (49.72%). Known pathogens were found significantly more often in cases of infantile wheeze compared to controls, exemplified by Haemophilus spp. (OR=2.12, 95% Confidence Interval (CI) 1.82-2.47; P=5.46×10(-23)) and Staphylococcus spp. (OR=124.1, 95%CI 59.0-261.2; P=1.87×10(-241)). Other OTUs were less common in cases than controls, notably Veillonella spp. (OR=0.59, 95%CI=0.56-0.62; P=8.06×10(-86)).
Discussion: The airway microbiota appeared to contain many more Streptococci than found in Western Europe and the USA. Comparisons between healthy and wheezing infants revealed a significant difference in several bacterial phylotypes that were not confounded by antibiotics or use of inhaled steroids. The increased prevalence of pathogens such as Haemophilus and Staphylococcus spp. in cases may contribute to wheezing illnesses in this age group.
Conflict of interest statement
Figures



References
-
- Cookson W (2004) The immunogenetics of asthma and eczema: a new focus on the epithelium. Nat Rev Immunol 4: 978–988. - PubMed
-
- Eder W, Ege MJ, von Mutius E (2006) The asthma epidemic. N Engl J Med 355: 2226–2235. - PubMed
-
- Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, et al. (2011) Exposure to environmental microorganisms and childhood asthma. N Engl J Med 364: 701–709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

