Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile
- PMID: 23071679
- PMCID: PMC3468616
- DOI: 10.1371/journal.pone.0046966
Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile
Abstract
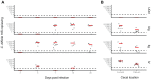
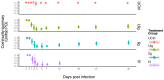
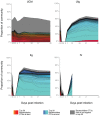
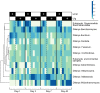
Antibiotic disruption of the intestinal microbiota may cause susceptibility to pathogens that is resolved by progressive bacterial outgrowth and colonization. Succession is central to ecological theory but not widely documented in studies of the vertebrate microbiome. Here, we study succession in the hamster gut after treatment with antibiotics and exposure to Clostridium difficile. C. difficile infection is typically lethal in hamsters, but protection can be conferred with neutralizing antibodies against the A and B toxins. We compare treatment with neutralizing monoclonal antibodies (mAb) to treatment with vancomycin, which prolongs the lives of animals but ultimately fails to protect them from death. We carried out longitudinal deep sequencing analysis and found distinctive waves of succession associated with each form of treatment. Clindamycin sensitization prior to infection was associated with the temporary suppression of the previously dominant Bacteroidales and the fungus Saccinobaculus in favor of Proteobacteria. In mAb-treated animals, C. difficile proliferated before joining Proteobacteria in giving way to re-expanding Bacteroidales and the fungus Wickerhamomyces. However, the Bacteroidales lineages returning by day 7 were different from those that were present initially, and they persisted for the duration of the experiment. Animals treated with vancomycin showed a different set of late-stage lineages that were dominated by Proteobacteria as well as increased disparity between the tissue-associated and luminal cecal communities. The control animals showed no change in their gut microbiota. These data thus suggest different patterns of ecological succession following antibiotic treatment and C. difficile infection.
Conflict of interest statement
Figures



References
-
- Armstrong TG, Wilmot AJ, Elsdon-Dew R (1950) Aureomycin and amoebic dysentery. Lancet 2: 10–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

