DNA polymerase α (swi7) and the flap endonuclease Fen1 (rad2) act together in the S-phase alkylation damage response in S. pombe
- PMID: 23071723
- PMCID: PMC3469492
- DOI: 10.1371/journal.pone.0047091
DNA polymerase α (swi7) and the flap endonuclease Fen1 (rad2) act together in the S-phase alkylation damage response in S. pombe
Abstract
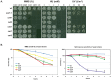
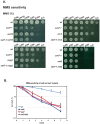
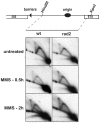
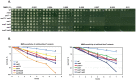
Polymerase α is an essential enzyme mainly mediating Okazaki fragment synthesis during lagging strand replication. A specific point mutation in Schizosaccharomyces pombe polymerase α named swi7-1, abolishes imprinting required for mating-type switching. Here we investigate whether this mutation confers any genome-wide defects. We show that the swi7-1 mutation renders cells hypersensitive to the DNA damaging agents methyl methansulfonate (MMS), hydroxyurea (HU) and UV and incapacitates activation of the intra-S checkpoint in response to DNA damage. In addition we show that, in the swi7-1 background, cells are characterized by an elevated level of repair foci and recombination, indicative of increased genetic instability. Furthermore, we detect novel Swi1-, -Swi3- and Pol α- dependent alkylation damage repair intermediates with mobility on 2D-gel that suggests presence of single-stranded regions. Genetic interaction studies showed that the flap endonuclease Fen1 works in the same pathway as Pol α in terms of alkylation damage response. Fen1 was also required for formation of alkylation- damage specific repair intermediates. We propose a model to explain how Pol α, Swi1, Swi3 and Fen1 might act together to detect and repair alkylation damage during S-phase.
Conflict of interest statement
Figures






Similar articles
-
Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage.Mol Cell Biol. 2005 Apr;25(7):2770-84. doi: 10.1128/MCB.25.7.2770-2784.2005. Mol Cell Biol. 2005. PMID: 15767681 Free PMC article.
-
Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex.J Biol Chem. 2005 Dec 30;280(52):42536-42. doi: 10.1074/jbc.M510575200. Epub 2005 Oct 31. J Biol Chem. 2005. PMID: 16263721
-
A conserved domain of Schizosaccharomyces pombe dfp1(+) is uniquely required for chromosome stability following alkylation damage during S phase.Mol Cell Biol. 2002 Jul;22(13):4477-90. doi: 10.1128/MCB.22.13.4477-4490.2002. Mol Cell Biol. 2002. PMID: 12052858 Free PMC article.
-
Repair of UV damage in the fission yeast Schizosaccharomyces pombe.Mutat Res. 2000 Jun 30;451(1-2):197-210. doi: 10.1016/s0027-5107(00)00050-6. Mutat Res. 2000. PMID: 10915873 Review.
-
S-phase DNA damage checkpoint in budding yeast.Biol Chem. 1998 Aug-Sep;379(8-9):1019-23. doi: 10.1515/bchm.1998.379.8-9.1019. Biol Chem. 1998. PMID: 9792433 Review.
Cited by
-
Characterization of a Novel MMS-Sensitive Allele of Schizosaccharomyces pombe mcm4.G3 (Bethesda). 2016 Oct 13;6(10):3049-3063. doi: 10.1534/g3.116.033571. G3 (Bethesda). 2016. PMID: 27473316 Free PMC article.
-
DNA replication machinery prevents Rad52-dependent single-strand annealing that leads to gross chromosomal rearrangements at centromeres.Commun Biol. 2020 Apr 30;3(1):202. doi: 10.1038/s42003-020-0934-0. Commun Biol. 2020. PMID: 32355220 Free PMC article.
-
The Fission Yeast Mating-Type Switching Motto: "One-for-Two" and "Two-for-One".Microbiol Mol Biol Rev. 2023 Mar 21;87(1):e0000821. doi: 10.1128/mmbr.00008-21. Epub 2023 Jan 11. Microbiol Mol Biol Rev. 2023. PMID: 36629411 Free PMC article. Review.
-
Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model.J Immunol. 2016 Aug 15;197(4):1477-88. doi: 10.4049/jimmunol.1600589. Epub 2016 Jul 1. J Immunol. 2016. PMID: 27371726 Free PMC article.
-
Role of swi7H4 mutant allele of DNA polymerase α in the DNA damage checkpoint response.PLoS One. 2015 Mar 30;10(3):e0124063. doi: 10.1371/journal.pone.0124063. eCollection 2015. PLoS One. 2015. PMID: 25822347 Free PMC article.
References
-
- Wang TS, Wong SW, Korn D (1989) Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. Faseb J 3: 14–21. - PubMed
-
- Singh J, Klar AJ (1993) DNA polymerase-alpha is essential for mating-type switching in fission yeast. Nature 361: 271–273. - PubMed
-
- Dalgaard JZ, Klar AJ (1999) Orientation of DNA replication establishes mating-type switching pattern in S. pombe . Nature 400: 181–184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

