Chromatin modification by PSC occurs at one PSC per nucleosome and does not require the acidic patch of histone H2A
- PMID: 23071745
- PMCID: PMC3469540
- DOI: 10.1371/journal.pone.0047162
Chromatin modification by PSC occurs at one PSC per nucleosome and does not require the acidic patch of histone H2A
Abstract
Chromatin architecture is regulated through both enzymatic and non-enzymatic activities. For example, the Polycomb Group (PcG) proteins maintain developmental gene silencing using an array of chromatin-based mechanisms. The essential Drosophila PcG protein, Posterior Sex Combs (PSC), compacts chromatin and inhibits chromatin remodeling and transcription through a non-enzymatic mechanism involving nucleosome bridging. Nucleosome bridging is achieved through a combination of nucleosome binding and self-interaction. Precisely how PSC interacts with chromatin to bridge nucleosomes is not known and is the subject of this work. We determine the stoichiometry of PSC-chromatin interactions in compact chromatin (in which nucleosomes are bridged) using Scanning Transmission Electron Microscopy (STEM). We find that full compaction occurs with one PSC per nucleosome. In addition to compacting chromatin, we show that PSC oligomerizes nucleosome arrays. PSC-mediated oligomerization of chromatin occurs at similar stoichiometry as compaction suggesting it may also involve nucleosome bridging. Interactions between the tail of histone H4 and the acidic patch of histone H2A are important for chromatin folding and oligomerization, and several chromatin proteins bind the histone H2A acidic patch. However, mutation of the acidic patch of histone H2A does not affect PSC's ability to inhibit chromatin remodeling or bridge nucleosomes. In fact, PSC does not require nucleosomes for bridging activity but can bridge naked DNA segments. PSC clusters nucleosomes on sparsely assembled templates, suggesting it interacts preferentially with nucleosomes over bare DNA. This may be due to the ability of PSC to bind free histones. Our data are consistent with a model in which each PSC binds a nucleosome and at least one other PSC to directly bridge nucleosomes and compact chromatin, but also suggest that naked DNA can be included in compacted structures. We discuss how our data highlight the diversity of mechanisms used to modify chromatin architecture.
Conflict of interest statement
Figures
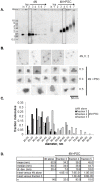
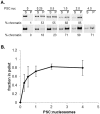
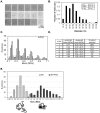

 .
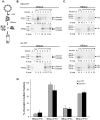
.


Similar articles
-
Inhibition of chromatin remodeling by polycomb group protein posterior sex combs is mechanistically distinct from nucleosome binding.Biochemistry. 2010 Nov 9;49(44):9438-48. doi: 10.1021/bi100532a. Biochemistry. 2010. PMID: 20873869 Free PMC article.
-
Regulation of Nucleosome Stacking and Chromatin Compaction by the Histone H4 N-Terminal Tail-H2A Acidic Patch Interaction.J Mol Biol. 2017 Jun 30;429(13):2075-2092. doi: 10.1016/j.jmb.2017.03.016. Epub 2017 Mar 16. J Mol Biol. 2017. PMID: 28322915
-
Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.Nature. 2018 Apr;556(7701):386-390. doi: 10.1038/s41586-018-0029-y. Epub 2018 Apr 11. Nature. 2018. PMID: 29643509 Free PMC article.
-
The role of the nucleosome acidic patch in modulating higher order chromatin structure.J R Soc Interface. 2013 Feb 27;10(82):20121022. doi: 10.1098/rsif.2012.1022. Print 2013 May 6. J R Soc Interface. 2013. PMID: 23446052 Free PMC article. Review.
-
All roads lead to chromatin: multiple pathways for histone deposition.Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):238-46. Biochim Biophys Acta. 2013. PMID: 24459726 Review.
Cited by
-
DNA Binding Reorganizes the Intrinsically Disordered C-Terminal Region of PSC in Drosophila PRC1.J Mol Biol. 2020 Aug 7;432(17):4856-4871. doi: 10.1016/j.jmb.2020.07.002. Epub 2020 Jul 3. J Mol Biol. 2020. PMID: 32628956 Free PMC article.
-
How is epigenetic information maintained through DNA replication?Epigenetics Chromatin. 2013 Oct 2;6(1):32. doi: 10.1186/1756-8935-6-32. Epigenetics Chromatin. 2013. PMID: 24225278 Free PMC article.
-
Distinct Cellular Assembly Stoichiometry of Polycomb Complexes on Chromatin Revealed by Single-molecule Chromatin Immunoprecipitation Imaging.J Biol Chem. 2015 Nov 20;290(47):28038-28054. doi: 10.1074/jbc.M115.671115. Epub 2015 Sep 17. J Biol Chem. 2015. PMID: 26381410 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

