Sequence requirements for localization and packaging of Ty3 retroelement RNA
- PMID: 23073180
- PMCID: PMC3578171
- DOI: 10.1016/j.virusres.2012.10.008
Sequence requirements for localization and packaging of Ty3 retroelement RNA
Abstract
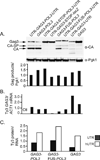
Retroviruses and retrotransposons package genomic RNA into virus-like particles (VLPs) in a poorly understood process. Expression of the budding yeast retrotransposon Ty3 results in the formation of cytoplasmic Ty3 VLP assembly foci comprised of Ty3 RNA and proteins, and cellular factors associated with RNA processing body (PB) components, which modulate translation and effect nonsense-mediated decay (NMD). A series of Ty3 RNA variants were tested to understand the effects of read-through translation via programmed frameshifting on RNA localization and packaging into VLPs, and to identify the roles of coding and non-coding sequences in those processes. These experiments showed that a low level of read-through translation of the downstream open reading frame (as opposed to no translation or translation without frameshifting) is important for localization of full-length Ty3 RNA to foci. Ty3 RNA variants associated with PB components via independent determinants in the native Ty3 untranslated regions (UTRs) and in GAG3-POL3 sequences flanked by UTRs adapted from non-Ty3 transcripts. However, despite localization, RNAs containing GAG3-POL3 but lacking Ty3 UTRs were not packaged efficiently. Surprisingly, sequences within Ty3 UTRs, which bind the initiator tRNA(Met) proposed to provide the dimerization interface, were not required for packaging of full-length Ty3 RNA into VLPs. In summary, our results demonstrate that Gag3 is sufficient and required for localization and packaging of RNAs containing Ty3 UTRs and support a role for POL3 sequences, translation of which is attenuated by programmed frameshifting, in both localization and packaging of the Ty3 full-length gRNA.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3.Viruses. 2016 Jul 14;8(7):193. doi: 10.3390/v8070193. Viruses. 2016. PMID: 27428991 Free PMC article. Review.
-
Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA.RNA Biol. 2019 Dec;16(12):1749-1763. doi: 10.1080/15476286.2019.1657370. Epub 2019 Aug 30. RNA Biol. 2019. PMID: 31469343 Free PMC article.
-
Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components.RNA. 2006 Jan;12(1):94-101. doi: 10.1261/rna.2264806. RNA. 2006. PMID: 16373495 Free PMC article.
-
Ty3, a Position-specific Retrotransposon in Budding Yeast.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0057-2014. doi: 10.1128/microbiolspec.MDNA3-0057-2014. Microbiol Spectr. 2015. PMID: 26104707 Review.
-
Function of a retrotransposon nucleocapsid protein.RNA Biol. 2010 Nov-Dec;7(6):642-54. doi: 10.4161/rna.7.6.14117. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21189452 Free PMC article. Review.
Cited by
-
A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag.Nucleic Acids Res. 2021 May 7;49(8):4668-4688. doi: 10.1093/nar/gkab223. Nucleic Acids Res. 2021. PMID: 33836091 Free PMC article.
-
Computational modeling of RNase, antisense ORF0 RNA, and intracellular compartmentation and their impact on the life cycle of the line retrotransposon.Comput Struct Biotechnol J. 2021 Oct 5;19:5667-5677. doi: 10.1016/j.csbj.2021.10.003. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34765087 Free PMC article.
-
Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3.Viruses. 2016 Jul 14;8(7):193. doi: 10.3390/v8070193. Viruses. 2016. PMID: 27428991 Free PMC article. Review.
-
Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA.RNA Biol. 2019 Dec;16(12):1749-1763. doi: 10.1080/15476286.2019.1657370. Epub 2019 Aug 30. RNA Biol. 2019. PMID: 31469343 Free PMC article.
-
Ty3 Retrotransposon Hijacks Mating Yeast RNA Processing Bodies to Infect New Genomes.PLoS Genet. 2015 Sep 30;11(9):e1005528. doi: 10.1371/journal.pgen.1005528. eCollection 2015. PLoS Genet. 2015. PMID: 26421679 Free PMC article.
References
-
- Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci. 2010;123:369–383. - PubMed
-
- Amberg D, BDSJD Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. 2005
-
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

