Molecular physiology of SPAK and OSR1: two Ste20-related protein kinases regulating ion transport
- PMID: 23073627
- PMCID: PMC4519243
- DOI: 10.1152/physrev.00009.2012
Molecular physiology of SPAK and OSR1: two Ste20-related protein kinases regulating ion transport
Abstract
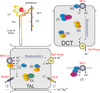
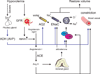
SPAK (Ste20-related proline alanine rich kinase) and OSR1 (oxidative stress responsive kinase) are members of the germinal center kinase VI subfamily of the mammalian Ste20 (Sterile20)-related protein kinase family. Although there are 30 enzymes in this protein kinase family, their conservation across the fungi, plant, and animal kingdom confirms their evolutionary importance. Already, a large volume of work has accumulated on the tissue distribution, binding partners, signaling cascades, and physiological roles of mammalian SPAK and OSR1 in multiple organ systems. After reviewing this basic information, we will examine newer studies that demonstrate the pathophysiological consequences to SPAK and/or OSR1 disruption, discuss the development and analysis of genetically engineered mouse models, and address the possible role these serine/threonine kinases might have in cancer proliferation and migration.
Figures









References
-
- Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101:2271–2290. - PubMed
-
- Adragna NC, Fulvio MD, Lauf PK. Regulation of K-Cl cotransport: from function to genes. J Membr Biol. 2004;201:109–137. - PubMed
-
- Alvarez-Leefmans FJ. Chloride transporters in presynaptic inhibition, pain and neurogenic inflammation. In: Alvarez-Leefmans FJ, Delpire E, editors. Physiology and Pathology of Chloride Transporter and Channels in the Nervous System: From molecules to diseases. London: Academic Press; 2009. pp. 439–470.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

