Putting the pieces together: high-performance LC-MS/MS provides network-, pathway-, and protein-level perspectives in Populus
- PMID: 23073815
- PMCID: PMC3536892
- DOI: 10.1074/mcp.M112.022996
Putting the pieces together: high-performance LC-MS/MS provides network-, pathway-, and protein-level perspectives in Populus
Abstract
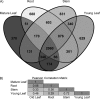
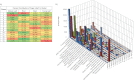
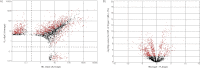
High-performance mass spectrometry (MS)-based proteomics enabled the construction of a detailed proteome atlas for Populus, a woody perennial plant model organism. Optimization of experimental procedures and implementation of current state-of-the-art instrumentation afforded the most detailed look into the predicted proteome space of Populus, offering varying proteome perspectives: (1) network-wide, (2) pathway-specific, and (3) protein-level viewpoints. Together, enhanced protein retrieval through a detergent-based lysis approach and maximized peptide sampling via the dual-pressure linear ion trap mass spectrometer (LTQ Velos), have resulted in the identification of 63,056 tryptic peptides. The technological advancements, specifically spectral-acquisition and sequencing speed, afforded the deepest look into the Populus proteome, with peptide abundances spanning 6 orders of magnitude and mapping to ∼25% of the predicted proteome space. In total, tryptic peptides mapped to 11,689 protein assignments across four organ-types: mature (fully expanded, leaf plastichronic index (LPI) 10-12) leaf, young (juvenile, LPI 4-6) leaf, root, and stem. To resolve protein ambiguity, identified proteins were grouped by sequence similarity (≥ 90%), thereby reducing the protein assignments into 7538 protein groups. In addition, this large-scale data set features the first systems-wide survey of protein expression across different Populus organs. As a demonstration of the precision and comprehensiveness of the semiquantitative analysis, we were able to contrast two stages of leaf development, mature versus young leaf. Statistical comparison through ANOVA analysis revealed 1432 protein groups that exhibited statistically significant (p ≤ 0.01) differences in protein abundance. Experimental validation of the metabolic circuitry expected in mature leaf (characterized by photosynthesis and carbon fixation) compared with young leaf (characterized by rapid growth and moderate photosynthetic activities) strongly testifies to the credibility of the approach. Instead of quantitatively comparing a few proteins, a systems view of all the changes associated with a given cellular perturbation could be made.
Figures




Similar articles
-
Analysis of the poplar phloem proteome and its response to leaf wounding.J Proteome Res. 2009 May;8(5):2341-50. doi: 10.1021/pr800968r. J Proteome Res. 2009. PMID: 19245218
-
Screening for changes in leaf and cambial proteome of Populus tremula × P. alba under different heat constraints.J Plant Physiol. 2012 Nov 15;169(17):1698-718. doi: 10.1016/j.jplph.2012.06.016. Epub 2012 Aug 9. J Plant Physiol. 2012. PMID: 22883629
-
Defining the boundaries and characterizing the landscape of functional genome expression in vascular tissues of Populus using shotgun proteomics.J Proteome Res. 2012 Jan 1;11(1):449-60. doi: 10.1021/pr200851y. Epub 2011 Nov 9. J Proteome Res. 2012. PMID: 22003893
-
Regulation of the leaf proteome by inoculation of Populus × canescens with two Paxillus involutus isolates differing in root colonization rates.Mycorrhiza. 2019 Oct;29(5):503-517. doi: 10.1007/s00572-019-00910-5. Epub 2019 Aug 27. Mycorrhiza. 2019. PMID: 31456074
-
[Recent progress in capillary electrophoresis-based high-sensitivity proteomics].Se Pu. 2020 Oct 8;38(10):1125-1132. doi: 10.3724/SP.J.1123.2020.03003. Se Pu. 2020. PMID: 34213109 Review. Chinese.
Cited by
-
In-depth assembly of organ and development dissected Picrorhiza kurroa proteome map using mass spectrometry.BMC Plant Biol. 2021 Dec 22;21(1):604. doi: 10.1186/s12870-021-03394-8. BMC Plant Biol. 2021. PMID: 34937558 Free PMC article.
-
Multi-Omics Methods Applied to Flower Development.Methods Mol Biol. 2023;2686:495-508. doi: 10.1007/978-1-0716-3299-4_23. Methods Mol Biol. 2023. PMID: 37540374
-
Epigenetic regulation of lignin biosynthesis in wood formation.New Phytol. 2025 Feb;245(4):1589-1607. doi: 10.1111/nph.20328. Epub 2024 Dec 5. New Phytol. 2025. PMID: 39639540 Free PMC article.
-
A PtrLBD39-mediated transcriptional network regulates tension wood formation in Populus trichocarpa.Plant Commun. 2021 Oct 20;3(1):100250. doi: 10.1016/j.xplc.2021.100250. eCollection 2022 Jan 10. Plant Commun. 2021. PMID: 35059630 Free PMC article.
-
Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis.J Proteome Res. 2014 Apr 4;13(4):1873-84. doi: 10.1021/pr400967x. Epub 2014 Mar 10. J Proteome Res. 2014. PMID: 24571389 Free PMC article.
References
-
- Ahrens C. H., Brunner E., Qeli E., Basler K., Aebersold R. (2010) Generating and navigating proteome maps using mass spectrometry. Nat. Rev. Mol. Cell Biol. 11, 789–801 - PubMed
-
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Schein J., Sterck L., Aerts A., Bhalerao R. R., Bhalerao R. P., Blaudez D., Boerjan W., Brun A., Brunner A., Busov V., Campbell M., Carlson J., Chalot M., Chapman J., Chen G. L., Cooper D., Coutinho P. M., Couturier J., Covert S., Cronk Q., Cunningham R., Davis J., Degroeve S., Dejardin A., Depamphilis C., Detter J., Dirks B., Dubchak I., Duplessis S., Ehlting J., Ellis B., Gendler K., Goodstein D., Gribskov M., Grimwood J., Groover A., Gunter L., Hamberger B., Heinze B., Helariutta Y., Henrissat B., Holligan D., Holt R., Huang W., Islam-Faridi N., Jones S., Jones-Rhoades M., Jorgensen R., Joshi C., Kangasjarvi J., Karlsson J., Kelleher C., Kirkpatrick R., Kirst M., Kohler A., Kalluri U., Larimer F., Leebens-Mack J., Leple J. C., Locascio P., Lou Y., Lucas S., Martin F., Montanini B., Napoli C., Nelson D. R., Nelson C., Nieminen K., Nilsson O., Pereda V., Peter G., Philippe R., Pilate G., Poliakov A., Razumovskaya J., Richardson P., Rinaldi C., Ritland K., Rouze P., Ryaboy D., Schmutz J., Schrader J., Segerman B., Shin H., Siddiqui A., Sterky F., Terry A., Tsai C. J., Uberbacher E., Unneberg P., Vahala J., Wall K., Wessler S., Yang G., Yin T., Douglas C., Marra M., Sandberg G., Van de Peer Y., Rokhsar D. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 - PubMed
-
- Plomion C., Lalanne C., Claverol S., Meddour H., Kohler A., Bogeat-Triboulot M. B., Barre A., Le Provost G., Dumazet H., Jacob D., Bastien C., Dreyer E., de Daruvar A., Guehl J. M., Schmitter J. M., Martin. F., Bonneu M. (2006) Mapping the proteome of poplar and application to the discovery of drought-stress responsive proteins. Proteomics 6, 6509–6527 - PubMed
-
- Kalluri U. C., Hurst G. B., Lankford P. K., Ranjan P., Pelletier D. A. (2009) Shotgun proteome profile of Populus developing xylem. Proteomics 9, 4871–4880 - PubMed
-
- Shuford C. M., Li Q., Sun Y. H., Chen H. C., Wang J., Shi R., Sederoff R. R., Chiang V. L., Muddiman D. C. (2012) Comprehensive quantification of monolignol-pathway enzymes in populus trichocarpa by protein cleavage isotope dilution mass spectrometry. J Proteome Res. 11, 3390–3404 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

