Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling
- PMID: 23074192
- PMCID: PMC3526270
- DOI: 10.1093/nar/gks956
Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling
Abstract
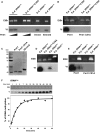
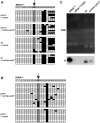
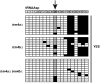
The fission yeast Schizosaccharomyces pombe carries a cytosine 5-methyltransferase homolog of the Dnmt2 family (termed pombe methyltransferase 1, Pmt1), but contains no detectable DNA methylation. Here, we found that Pmt1, like other Dnmt2 homologs, has in vitro methylation activity on cytosine 38 of tRNA(Asp) and, to a lesser extent, of tRNA(Glu), despite the fact that it contains a non-consensus residue in catalytic motif IV as compared with its homologs. In vivo tRNA methylation also required Pmt1. Unexpectedly, however, its in vivo activity showed a strong dependence on the nutritional status of the cell because Pmt1-dependent tRNA methylation was induced in cells grown in the presence of peptone or with glutamate as a nitrogen source. Furthermore, this induction required the serine/threonine kinase Sck2, but not the kinases Sck1, Pka1 or Tor1 and was independent of glucose signaling. Taken together, this work reveals a novel connection between nutrient signaling and tRNA methylation that thus may link tRNA methylation to processes downstream of nutrient signaling like ribosome biogenesis and translation initiation.
Figures


References
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. - PubMed
-
- Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011;2:611–631. - PubMed
-
- Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. - PubMed
-
- Hermann A, Schmitt S, Jeltsch A. The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem. 2003;278:31717–31721. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

