Efficient detection, quantification and enrichment of subtle allelic alterations
- PMID: 23075543
- PMCID: PMC3473374
- DOI: 10.1093/dnares/dss023
Efficient detection, quantification and enrichment of subtle allelic alterations
Abstract
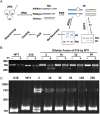
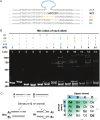
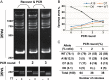
Gene targeting (GT) can introduce subtle alterations into a particular locus and represents a powerful tool for genome editing. Engineered zinc finger nucleases (ZFNs) are effective for generating minor allelic alterations. Efficient detection of such minor alterations remains one of the challenges in ZFN-mediated GT experiments. Here, we report the establishment of procedures allowing for efficient detection, quantification and enrichment of such subtle alterations. In a biallelic model, polyacrylamide gel electrophoresis (PAGE) is capable of detecting rare allelic variations in the form of DNA heteroduplexes at a high efficiency of ~0.4% compared with ~6.3% by the traditional T7 endonuclease I-digestion and agarose gel electrophoresis. In a multiple allelic model, PAGE could discriminate different alleles bearing addition or deletion of 1-18 bp as distinct bands that were easily quantifiable by densitometry. Furthermore, PAGE enables enrichment for rare alleles. We show for the first time that direct endogenous GT is possible in medaka by ZFN RNA injection, whereas PAGE allows for detection and cloning of ZFN-targeted alleles in adults arising from ZFN-injected medaka embryos. Therefore, PAGE is effective for detection, quantification and enrichment of multiple fine allelic differences and thus offers a versatile tool for screening targeted subtle gene alterations.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

