Drift-barrier hypothesis and mutation-rate evolution
- PMID: 23077252
- PMCID: PMC3494944
- DOI: 10.1073/pnas.1216223109
Drift-barrier hypothesis and mutation-rate evolution
Abstract
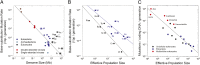
Mutation dictates the tempo and mode of evolution, and like all traits, the mutation rate is subject to evolutionary modification. Here, we report refined estimates of the mutation rate for a prokaryote with an exceptionally small genome and for a unicellular eukaryote with a large genome. Combined with prior results, these estimates provide the basis for a potentially unifying explanation for the wide range in mutation rates that exists among organisms. Natural selection appears to reduce the mutation rate of a species to a level that scales negatively with both the effective population size (N(e)), which imposes a drift barrier to the evolution of molecular refinements, and the genomic content of coding DNA, which is proportional to the target size for deleterious mutations. As a consequence of an expansion in genome size, some microbial eukaryotes with large N(e) appear to have evolved mutation rates that are lower than those known to occur in prokaryotes, but multicellular eukaryotes have experienced elevations in the genome-wide deleterious mutation rate because of substantial reductions in N(e).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Proteome size as the major factor determining mutation rates.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):E858-9. doi: 10.1073/pnas.1219306110. Epub 2013 Feb 13. Proc Natl Acad Sci U S A. 2013. PMID: 23407172 Free PMC article. No abstract available.
-
Reply to Massey: Drift does influence mutation-rate evolution.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):E860. doi: 10.1073/pnas.1220650110. Proc Natl Acad Sci U S A. 2013. PMID: 23593647 Free PMC article. No abstract available.
References
-
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17(10):589–596. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

