mRNA decay during herpes simplex virus (HSV) infections: mutations that affect translation of an mRNA influence the sites at which it is cleaved by the HSV virion host shutoff (Vhs) protein
- PMID: 23077305
- PMCID: PMC3536374
- DOI: 10.1128/JVI.01557-12
mRNA decay during herpes simplex virus (HSV) infections: mutations that affect translation of an mRNA influence the sites at which it is cleaved by the HSV virion host shutoff (Vhs) protein
Abstract
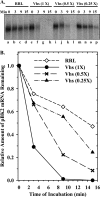
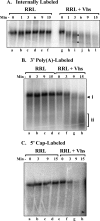
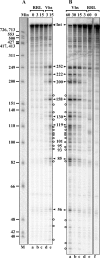
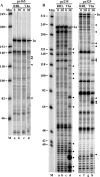
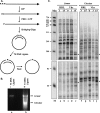
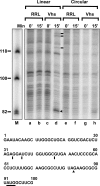
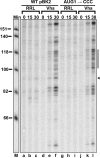
During lytic infections, the herpes simplex virus (HSV) virion host shutoff (Vhs) endoribonuclease degrades many host and viral mRNAs. Within infected cells it cuts mRNAs at preferred sites, including some in regions of translation initiation. Vhs binds the translation initiation factors eIF4H, eIF4AI, and eIF4AII, suggesting that its mRNA degradative function is somehow linked to translation. To explore how Vhs is targeted to preferred sites, we examined the in vitro degradation of a target mRNA in rabbit reticulocyte lysates containing in vitro-translated Vhs. Vhs caused rapid degradation of mRNAs beginning with cleavages at sites in the first 250 nucleotides, including a number near the start codon and in the 5' untranslated region. Ligation of the ends to form a circular mRNA inhibited Vhs cleavage at the same sites at which it cuts capped linear molecules. This was not due to an inability to cut any circular RNA, since Vhs cuts circular mRNAs containing an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) at the same sites as linear molecules with the IRES. Cutting linear mRNAs at preferred sites was augmented by the presence of a 5' cap. Moreover, mutations that altered the 5' proximal AUG abolished Vhs cleavage at nearby sites, while mutations that changed sequences surrounding the AUG to improve their match to the Kozak consensus sequence enhanced Vhs cutting near the start codon. The results indicate that mutations in an mRNA that affect its translation affect the sites at which it is cut by Vhs and suggest that Vhs is directed to its preferred cut sites during translation initiation.
Figures






References
-
- Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex viruses, p 2501–2601 In Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA
-
- Sandri-Goldin RM. 2007. Initiation of transcription and RNA synthesis, processing and transport in HSV and VZV infected cells. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: http://www.ncbi.nlm.nih.gov/books/NBK47363/ - PubMed
-
- Kristie TM. 2007. Early events pre-initiation of alphaherpes viral gene expression. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: http://www.ncbi.nlm.nih.gov/books/NBK47386/ - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

