Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions
- PMID: 23078205
- PMCID: PMC3524407
- DOI: 10.1111/mmi.12071
Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions
Abstract
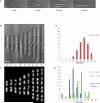
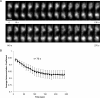
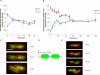
We examine whether the Escherichia coli chromosome is folded into a self-adherent nucleoprotein complex, or alternately is a confined but otherwise unconstrained self-avoiding polymer. We address this through in vivo visualization, using an inducible GFP fusion to the nucleoid-associated protein Fis to non-specifically decorate the entire chromosome. For a range of different growth conditions, the chromosome is a compact structure that does not fill the volume of the cell, and which moves from the new pole to the cell centre. During rapid growth, chromosome segregation occurs well before cell division, with daughter chromosomes coupled by a thin inter-daughter filament before complete segregation, whereas during slow growth chromosomes stay adjacent until cell division occurs. Image correlation analysis indicates that sub-nucleoid structure is stable on a 1 min timescale, comparable to the timescale for redistribution time measured for GFP-Fis after photobleaching. Optical deconvolution and writhe calculation analysis indicate that the nucleoid has a large-scale coiled organization rather than being an amorphous mass. Our observations are consistent with the chromosome having a self-adherent filament organization.
© 2012 Blackwell Publishing Ltd.
Figures



Comment in
-
Let's get 'Fisical' with bacterial nucleoid.Mol Microbiol. 2012 Dec;86(6):1285-90. doi: 10.1111/mmi.12073. Epub 2012 Oct 29. Mol Microbiol. 2012. PMID: 23078263
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

