A single nucleotide polymorphism in IL28B affects viral evolution of hepatitis C quasispecies after pegylated interferon and ribavirin therapy
- PMID: 23080496
- PMCID: PMC3481197
- DOI: 10.1002/jmv.23407
A single nucleotide polymorphism in IL28B affects viral evolution of hepatitis C quasispecies after pegylated interferon and ribavirin therapy
Abstract
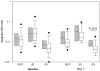
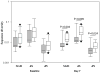
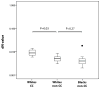
Interleukin-28B (IL28B) polymorphisms are associated with viral response to peginterferon and ribavirin (RBV) in chronic hepatitis C (HCV). Their recognition represents a breakthrough in the understanding of the role of the host in viral eradication. How these polymorphisms determine viral eradication is unknown. The IL-28B variants are hypothesized to have a differential impact on HCV quasispecies evolution during treatment with pegylated interferon (PEG-IFN) and RBV. In this study, HCV RNA levels were measured at early time points in 33 naïve genotype 1 hepatitis C patients and clonal analysis of the entire NS5A region was performed on sera from baseline and Day 7. Site rs12979860 polymorphisms were determined by direct sequencing of PCR products and classified into CC, CT, and TT and were identified in 13, 11, and 9 patients, respectively. The CC polymorphism more commonly was seen in Whites versus Blacks [12/21 (57%) vs. 1/12 (8%), P = 0.009] and HIV-infected versus mono-infected [13/25 (52%) vs. 0/8 (0%), P = 0.009]. Patients with CC and non-CC had similar baseline viral loads. More patients with the CC polymorphism had amino acid substitutions in NS5A compared to non-CC patients. Despite similar baseline viral diversity, by Day 7, significantly more patients with CC had higher non-synonymous substitution values compared to non-CC (P = 0.02). Chronic hepatitis C patients with the CC IL28B polymorphism have a higher number of amino acid substitutions in the NS5A region and early viral evolution due to greater interferon induced selective pressure during this critical period of treatment.
Copyright © 2012 Wiley Periodicals, Inc.
Conflict of interest statement
Potential conflict of interest: MKJ is a consultant for Merck and has been on the advisory board for Vertex and Boehringer Ingelheim. She has received research grant support from Vertex, Pfizer, ViiV Healthcare, Boehringer Ingelheim, Roche, Tibotec, Pfizer, Bristol-Myers Squibb, and Gilead. WML has been a consultant for Eli Lilly, Merck, Novartis, Pfizer, FoldRx and Gilead Sciences. WML has received research grant support from Schering-Plough Research Institute, Siemens, Vertex, Roche, Anadys, Gilead Sciences and Globeimmune.
Figures

References
-
- Chayama K, Hayes CN. Hepatitis C virus: How genetic variability affects pathobiology of disease. J Gastroenterol Hepatol. 2011;26(Suppl 1):83–95. - PubMed
-
- Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. - PubMed
-
- Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. - PubMed
-
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. - PubMed
-
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;16:16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

