The genome sequence of Propionibacterium acidipropionici provides insights into its biotechnological and industrial potential
- PMID: 23083487
- PMCID: PMC3534718
- DOI: 10.1186/1471-2164-13-562
The genome sequence of Propionibacterium acidipropionici provides insights into its biotechnological and industrial potential
Abstract
Background: Synthetic biology allows the development of new biochemical pathways for the production of chemicals from renewable sources. One major challenge is the identification of suitable microorganisms to hold these pathways with sufficient robustness and high yield. In this work we analyzed the genome of the propionic acid producer Actinobacteria Propionibacterium acidipropionici (ATCC 4875).
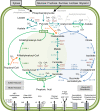
Results: The assembled P. acidipropionici genome has 3,656,170 base pairs (bp) with 68.8% G + C content and a low-copy plasmid of 6,868 bp. We identified 3,336 protein coding genes, approximately 1000 more than P. freudenreichii and P. acnes, with an increase in the number of genes putatively involved in maintenance of genome integrity, as well as the presence of an invertase and genes putatively involved in carbon catabolite repression. In addition, we made an experimental confirmation of the ability of P. acidipropionici to fix CO2, but no phosphoenolpyruvate carboxylase coding gene was found in the genome. Instead, we identified the pyruvate carboxylase gene and confirmed the presence of the corresponding enzyme in proteome analysis as a potential candidate for this activity. Similarly, the phosphate acetyltransferase and acetate kinase genes, which are considered responsible for acetate formation, were not present in the genome. In P. acidipropionici, a similar function seems to be performed by an ADP forming acetate-CoA ligase gene and its corresponding enzyme was confirmed in the proteome analysis.
Conclusions: Our data shows that P. acidipropionici has several of the desired features that are required to become a platform for the production of chemical commodities: multiple pathways for efficient feedstock utilization, ability to fix CO2, robustness, and efficient production of propionic acid, a potential precursor for valuable 3-carbon compounds.
Figures





Similar articles
-
Effects of carbon dioxide on cell growth and propionic acid production from glycerol and glucose by Propionibacterium acidipropionici.Bioresour Technol. 2015 Jan;175:374-81. doi: 10.1016/j.biortech.2014.10.046. Epub 2014 Oct 22. Bioresour Technol. 2015. PMID: 25459845
-
Understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the level of proteomics.Sci Rep. 2014 Nov 7;4:6951. doi: 10.1038/srep06951. Sci Rep. 2014. PMID: 25377721 Free PMC article.
-
Development of a Propionibacterium-Escherichia coli shuttle vector for metabolic engineering of Propionibacterium jensenii, an efficient producer of propionic acid.Appl Environ Microbiol. 2013 Aug;79(15):4595-602. doi: 10.1128/AEM.00737-13. Epub 2013 May 24. Appl Environ Microbiol. 2013. PMID: 23709505 Free PMC article.
-
Engineering propionibacteria as versatile cell factories for the production of industrially important chemicals: advances, challenges, and prospects.Appl Microbiol Biotechnol. 2015 Jan;99(2):585-600. doi: 10.1007/s00253-014-6228-z. Epub 2014 Nov 28. Appl Microbiol Biotechnol. 2015. PMID: 25431012 Review.
-
Biosynthesis of some organic acids and lipids in industrially important microorganisms is promoted by pyruvate carboxylases.J Biosci. 2019 Jun;44(2):47. J Biosci. 2019. PMID: 31180060 Review.
Cited by
-
Microbial and Genetic Resources for Cobalamin (Vitamin B12) Biosynthesis: From Ecosystems to Industrial Biotechnology.Int J Mol Sci. 2021 Apr 26;22(9):4522. doi: 10.3390/ijms22094522. Int J Mol Sci. 2021. PMID: 33926061 Free PMC article. Review.
-
Genome Sequence of Propionibacterium acidipropionici ATCC 55737.Genome Announc. 2016 May 19;4(3):e00248-16. doi: 10.1128/genomeA.00248-16. Genome Announc. 2016. PMID: 27198010 Free PMC article.
-
Steering the product spectrum in high-pressure anaerobic processes: CO2 partial pressure as a novel tool in biorefinery concepts.Biotechnol Biofuels Bioprod. 2023 Feb 18;16(1):27. doi: 10.1186/s13068-023-02262-x. Biotechnol Biofuels Bioprod. 2023. PMID: 36803622 Free PMC article.
-
Development of an industrializable fermentation process for propionic acid production.J Ind Microbiol Biotechnol. 2014 May;41(5):837-52. doi: 10.1007/s10295-014-1423-6. Epub 2014 Mar 14. J Ind Microbiol Biotechnol. 2014. PMID: 24627047
-
Propionibacterium freudenreichii thrives in microaerobic conditions by complete oxidation of lactate to CO2.Environ Microbiol. 2021 Jun;23(6):3116-3129. doi: 10.1111/1462-2920.15532. Epub 2021 May 6. Environ Microbiol. 2021. PMID: 33955639 Free PMC article.
References
-
- Rincones J, Zeidler AF, Grassi MCB, Carazzolle MF, Pereira GAG. The golden bridge for nature: the New biology applied to Bioplastics. Polym Rev. 2009;49:85–106. doi: 10.1080/15583720902834817. - DOI
-
- Zhu Y, Li J, Tan M, Liu L, Jiang L, Sun J, Lee P, Du G, Chen J. Optimization and scale-up of propionic acid production by propionic acid-tolerant Propionibacterium acidipropionici with glycerol as the carbon source. Bioresour Technol. 2010;101:8902–8906. doi: 10.1016/j.biortech.2010.06.070. - DOI - PubMed
-
- Boyaval P, Corre C. Production of propionic acid. Lait. 1995;75:453–461. doi: 10.1051/lait:19954-535. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

