Structure and orientation of bovine lactoferrampin in the mimetic bacterial membrane as revealed by solid-state NMR and molecular dynamics simulation
- PMID: 23083717
- PMCID: PMC3475389
- DOI: 10.1016/j.bpj.2012.09.010
Structure and orientation of bovine lactoferrampin in the mimetic bacterial membrane as revealed by solid-state NMR and molecular dynamics simulation
Abstract
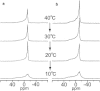
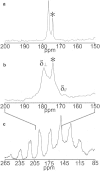
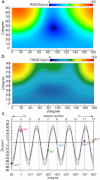
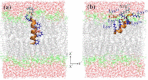
Bovine lactoferrampin (LFampinB) is a newly discovered antimicrobial peptide found in the N1-domain of bovine lactoferrin (268-284), and consists of 17 amino-acid residues. It is important to determine the orientation and structure of LFampinB in bacterial membranes to reveal the antimicrobial mechanism. We therefore performed (13)C and (31)P NMR, (13)C-(31)P rotational echo double resonance (REDOR), potassium ion-selective electrode, and quartz-crystal microbalance measurements for LFampinB with mimetic bacterial membrane and molecular-dynamics simulation in acidic membrane. (31)P NMR results indicated that LFampinB caused a defect in mimetic bacterial membranes. Ion-selective electrode measurements showed that ion leakage occurred for the mimetic bacterial membrane containing cardiolipin. Quartz-crystal microbalance measurements revealed that LFampinB had greater affinity to acidic phospholipids than that to neutral phospholipids. (13)C DD-MAS and static NMR spectra showed that LFampinB formed an α-helix in the N-terminus region and tilted 45° to the bilayer normal. REDOR dephasing patterns between carbonyl carbon nucleus in LFampinB and phosphorus nuclei in lipid phosphate groups were measured by (13)C-(31)P REDOR and the results revealed that LFampinB is located in the interfacial region of the membrane. Molecular-dynamics simulation showed the tilt angle to be 42° and the rotation angle to be 92.5° for Leu(3), which are in excellent agreement with the experimental values.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Solution structures and model membrane interactions of lactoferrampin, an antimicrobial peptide derived from bovine lactoferrin.Biochim Biophys Acta. 2007 Oct;1768(10):2355-64. doi: 10.1016/j.bbamem.2007.04.018. Epub 2007 May 1. Biochim Biophys Acta. 2007. PMID: 17560539
-
Interactions of bovine lactoferricin with acidic phospholipid bilayers and its antimicrobial activity as studied by solid-state NMR.Biochim Biophys Acta. 2006 Sep;1758(9):1523-8. doi: 10.1016/j.bbamem.2006.06.014. Epub 2006 Jun 18. Biochim Biophys Acta. 2006. PMID: 16884683
-
Influence of specific amino acid side-chains on the antimicrobial activity and structure of bovine lactoferrampin.Biochem Cell Biol. 2012 Jun;90(3):362-77. doi: 10.1139/o11-057. Epub 2012 Jan 17. Biochem Cell Biol. 2012. PMID: 22250712
-
Interaction of extracellular loop II of κ-opioid receptor (196-228) with opioid peptide dynorphin in membrane environments as revealed by solid state nuclear magnetic resonance, quartz crystal microbalance and molecular dynamics simulation.J Phys Chem B. 2014 Aug 14;118(32):9604-12. doi: 10.1021/jp505412j. Epub 2014 Jul 31. J Phys Chem B. 2014. PMID: 25059685
-
Structure and dynamics of phospholipids in membranes elucidated by combined use of NMR and vibrational spectroscopies.Biochim Biophys Acta Biomembr. 2020 Sep 1;1862(9):183352. doi: 10.1016/j.bbamem.2020.183352. Epub 2020 May 11. Biochim Biophys Acta Biomembr. 2020. PMID: 32407775 Review.
Cited by
-
Mechanisms of Action for Antimicrobial Peptides With Antibacterial and Antibiofilm Functions.Front Microbiol. 2019 Dec 12;10:2866. doi: 10.3389/fmicb.2019.02866. eCollection 2019. Front Microbiol. 2019. PMID: 31921046 Free PMC article. Review.
-
Novel lactotransferrin-derived synthetic peptides suppress cariogenic bacteria in vitro and arrest dental caries in vivo: [Novel lactotransferrin-derived anticaries peptides].J Oral Microbiol. 2021 Jun 20;13(1):1943999. doi: 10.1080/20002297.2021.1943999. eCollection 2021. J Oral Microbiol. 2021. PMID: 34234894 Free PMC article.
-
Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein.Int J Pept. 2013;2013:390230. doi: 10.1155/2013/390230. Epub 2013 Feb 13. Int J Pept. 2013. PMID: 23554820 Free PMC article.
-
Lipid composition-dependent membrane fragmentation and pore-forming mechanisms of membrane disruption by pexiganan (MSI-78).Biochemistry. 2013 May 14;52(19):3254-63. doi: 10.1021/bi400087n. Epub 2013 Apr 29. Biochemistry. 2013. PMID: 23590672 Free PMC article.
-
Three Decades of REDOR in Protein Science: A Solid-State NMR Technique for Distance Measurement and Spectral Editing.Int J Mol Sci. 2023 Sep 4;24(17):13637. doi: 10.3390/ijms241713637. Int J Mol Sci. 2023. PMID: 37686450 Free PMC article. Review.
References
-
- van der Kraan M.I.A., Groenink J., Nieuw Amerongen A.V. Lactoferrampin: a novel antimicrobial peptide in the N1-domain of bovine lactoferrin. Peptides. 2004;25:177–183. - PubMed
-
- van der Kraan M.I.A., van Marle J., Nieuw Amerongen A.V. Ultrastructural effects of antimicrobial peptides from bovine lactoferrin on the membranes of Candida albicans and Escherichia coli. Peptides. 2005;26:1537–1542. - PubMed
-
- van der Kraan M.I.A., Nazmi K., Nieuw Amerongen A.V. Lactoferrampin, an antimicrobial peptide of bovine lactoferrin, exerts its candidacidal activity by a cluster of positively charged residues at the C-terminus in combination with a helix-facilitating N-terminal part. Biol. Chem. 2005;386:137–142. - PubMed
-
- Haney E.F., Lau F., Vogel H.J. Solution structures and model membrane interactions of lactoferrampin, an antimicrobial peptide derived from bovine lactoferrin. Biochim. Biophys. Acta. 2007;1768:2355–2364. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

