Lack of CAK complex accumulation at DNA damage sites in XP-B and XP-B/CS fibroblasts reveals differential regulation of CAK anchoring to core TFIIH by XPB and XPD helicases during nucleotide excision repair
- PMID: 23083890
- PMCID: PMC3501591
- DOI: 10.1016/j.dnarep.2012.09.003
Lack of CAK complex accumulation at DNA damage sites in XP-B and XP-B/CS fibroblasts reveals differential regulation of CAK anchoring to core TFIIH by XPB and XPD helicases during nucleotide excision repair
Abstract
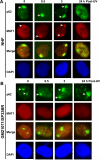
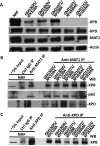
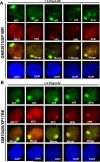
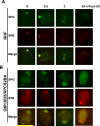
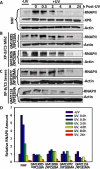
Transcription factor II H (TFIIH) is composed of core TFIIH and Cdk-activating kinase (CAK) complexes. Besides transcription, TFIIH also participates in nucleotide excision repair (NER), verifying DNA lesions through its helicase components XPB and XPD. The assembly state of TFIIH is known to be affected by truncation mutations in xeroderma pigmentosum group G/Cockayne syndrome (XP-G/CS). Here, we showed that CAK component MAT1 was rapidly recruited to UV-induced DNA damage sites, co-localizing with core TFIIH component p62, and dispersed from the damage sites upon completion of DNA repair. While the core TFIIH-CAK association remained intact, MAT1 failed to accumulate at DNA damage sites in fibroblasts harboring XP-B or XP-B/CS mutations. Nevertheless, MAT1, XPD and XPC as well as XPG were able to accumulate at damage sites in XP-D fibroblasts, in which the core TFIIH-CAK association also remained intact. Interestingly, XPG recruitment was impaired in XP-B/CS fibroblasts derived from patients with mild phenotype, but persisted in XP-B/CS fibroblasts from severely affected patients resulting in a nonfunctional preincision complex. An examination of steady-state levels of RNA polymerase II (RNAPII) indicated that UV-induced RNAPII phosphorylation was dramatically reduced in XP-B/CS fibroblasts. These results demonstrated that the CAK rapidly disassociates from the core TFIIH upon assembly of nonfunctional preincision complex in XP-B and XP-B/CS cells. The persistency of nonfunctional preincision complex correlates with the severity exhibited by XP-B patients. The results suggest that XPB and XPD helicases differentially regulate the anchoring of CAK to core TFIIH during damage verification step of NER.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase.DNA Repair (Amst). 2011 Jul 15;10(7):697-713. doi: 10.1016/j.dnarep.2011.04.028. Epub 2011 May 14. DNA Repair (Amst). 2011. PMID: 21571596 Free PMC article. Review.
-
Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy.Hum Mutat. 2008 Oct;29(10):1194-208. doi: 10.1002/humu.20768. Hum Mutat. 2008. PMID: 18470933 Free PMC article.
-
Dissociation of CAK from core TFIIH reveals a functional link between XP-G/CS and the TFIIH disassembly state.PLoS One. 2010 Jun 8;5(6):e11007. doi: 10.1371/journal.pone.0011007. PLoS One. 2010. PMID: 20543986 Free PMC article.
-
Influence of XPB helicase on recruitment and redistribution of nucleotide excision repair proteins at sites of UV-induced DNA damage.DNA Repair (Amst). 2007 Sep 1;6(9):1359-70. doi: 10.1016/j.dnarep.2007.03.025. Epub 2007 May 16. DNA Repair (Amst). 2007. PMID: 17509950 Free PMC article.
-
Hot topics in DNA repair: the molecular basis for different disease states caused by mutations in TFIIH and XPG.DNA Repair (Amst). 2008 Feb 1;7(2):339-44. doi: 10.1016/j.dnarep.2007.10.007. DNA Repair (Amst). 2008. PMID: 18077223 Free PMC article. Review.
Cited by
-
USP3 counteracts RNF168 via deubiquitinating H2A and γH2AX at lysine 13 and 15.Cell Cycle. 2014;13(1):106-14. doi: 10.4161/cc.26814. Epub 2013 Oct 24. Cell Cycle. 2014. PMID: 24196443 Free PMC article.
-
Timely upstream events regulating nucleotide excision repair by ubiquitin-proteasome system: ubiquitin guides the way.DNA Repair (Amst). 2021 Jul;103:103128. doi: 10.1016/j.dnarep.2021.103128. Epub 2021 May 12. DNA Repair (Amst). 2021. PMID: 33991872 Free PMC article. Review.
-
Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.Nat Struct Mol Biol. 2019 Jun;26(6):397-406. doi: 10.1038/s41594-019-0220-3. Epub 2019 May 20. Nat Struct Mol Biol. 2019. PMID: 31110295 Free PMC article.
-
The in vitro Analysis of Quality of Ovarian Follicle Culture Systems Using Time-Lapse Microscopy and Quantitative Real-Time PCR.J Reprod Infertil. 2020 Apr-Jun;21(2):94-106. J Reprod Infertil. 2020. PMID: 32500012 Free PMC article.
-
USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168.Cell Cycle. 2015;14(9):1413-25. doi: 10.1080/15384101.2015.1007785. Cell Cycle. 2015. PMID: 25894431 Free PMC article.
References
-
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. - PubMed
-
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. - PubMed
-
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. - PubMed
-
- Petit C, Sancar A. Nucleotide excision repair: From E.coli to man. Biochimie. 1999;81:15–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

