Contribution of SUMO-interacting motifs and SUMOylation to the antiretroviral properties of TRIM5α
- PMID: 23084420
- PMCID: PMC3534947
- DOI: 10.1016/j.virol.2012.09.042
Contribution of SUMO-interacting motifs and SUMOylation to the antiretroviral properties of TRIM5α
Abstract
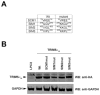
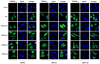
Recent findings suggested that the SUMO-interacting motifs (SIMs) present in the human TRIM5α (TRIM5α(hu)) protein play an important role in the ability of TRIM5α(hu) to restrict N-MLV. Here we explored the role of SIMs in the ability of rhesus TRIM5α (TRIM5α(rh)) to restrict HIV-1, and found that TRIM5α(rh) SIM mutants IL376KK (SIM1mut) and VI405KK (SIM2mut) completely lost their ability to block HIV-1 infection. Interestingly, these mutants also lost the recently described property of TRIM5α(rh) to shuttle into the nucleus. Analysis of these variants revealed that they are unable to interact with the HIV-1 core, which might explain the reason that these variants are not active against HIV-1. Furthermore, NMR titration experiments to assay the binding between the PRYSPRY domain of TRIM5α(rh) and the small ubiquitin-like modifier 1(SUMO-1) revealed no interaction. In addition, we examined the role of SUMOylation in restriction, and find out that inhibition of SUMOylation by the adenoviral protein Gam1 did not alter the retroviral restriction ability of TRIM5α. Overall, our results do not support a role for SIMs or SUMOylation in the antiviral properties of TRIM5α.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


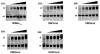




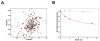
Similar articles
-
TRIM5α is a SUMO substrate.Retrovirology. 2015 Mar 24;12:28. doi: 10.1186/s12977-015-0155-7. Retrovirology. 2015. PMID: 25880753 Free PMC article.
-
SUMO-interacting motifs of human TRIM5α are important for antiviral activity.PLoS Pathog. 2011 Apr;7(4):e1002019. doi: 10.1371/journal.ppat.1002019. Epub 2011 Apr 7. PLoS Pathog. 2011. PMID: 21490953 Free PMC article.
-
Role of SUMO-1 and SUMO interacting motifs in rhesus TRIM5α-mediated restriction.Retrovirology. 2013 Feb 1;10:10. doi: 10.1186/1742-4690-10-10. Retrovirology. 2013. PMID: 23369348 Free PMC article.
-
Control of nuclear activities by substrate-selective and protein-group SUMOylation.Annu Rev Genet. 2013;47:167-86. doi: 10.1146/annurev-genet-111212-133453. Epub 2013 Aug 30. Annu Rev Genet. 2013. PMID: 24016193 Review.
-
TRIM5alpha.Curr Top Microbiol Immunol. 2009;339:47-66. doi: 10.1007/978-3-642-02175-6_3. Curr Top Microbiol Immunol. 2009. PMID: 20012523 Review.
Cited by
-
TRIM5α is a SUMO substrate.Retrovirology. 2015 Mar 24;12:28. doi: 10.1186/s12977-015-0155-7. Retrovirology. 2015. PMID: 25880753 Free PMC article.
-
TRIM5α Promotes Ubiquitination of Rta from Epstein-Barr Virus to Attenuate Lytic Progression.Front Microbiol. 2017 Jan 5;7:2129. doi: 10.3389/fmicb.2016.02129. eCollection 2016. Front Microbiol. 2017. PMID: 28105027 Free PMC article.
-
Endogenous TRIM5α Function Is Regulated by SUMOylation and Nuclear Sequestration for Efficient Innate Sensing in Dendritic Cells.Cell Rep. 2016 Jan 12;14(2):355-69. doi: 10.1016/j.celrep.2015.12.039. Epub 2015 Dec 31. Cell Rep. 2016. PMID: 26748714 Free PMC article.
-
Interplay between viruses and host sumoylation pathways.Nat Rev Microbiol. 2013 Jun;11(6):400-11. doi: 10.1038/nrmicro3015. Epub 2013 Apr 29. Nat Rev Microbiol. 2013. PMID: 23624814 Review.
-
The innate immune roles of host factors TRIM5α and Cyclophilin A on HIV-1 replication.Med Microbiol Immunol. 2015 Oct;204(5):557-65. doi: 10.1007/s00430-015-0417-y. Epub 2015 Apr 19. Med Microbiol Immunol. 2015. PMID: 25894765 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

