An in vivo functional screen uncovers miR-150-mediated regulation of hematopoietic injury response
- PMID: 23084747
- PMCID: PMC3487471
- DOI: 10.1016/j.celrep.2012.09.014
An in vivo functional screen uncovers miR-150-mediated regulation of hematopoietic injury response
Abstract
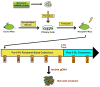
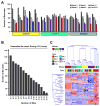
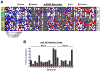
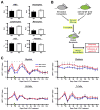
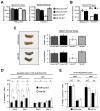
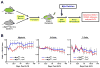
Hematopoietic stem and progenitor cells are often undesired targets of chemotherapies, leading to hematopoietic suppression requiring careful clinical management. Whether microRNAs control hematopoietic injury response is largely unknown. We report an in vivo gain-of-function screen and the identification of miR-150 as an inhibitor of hematopoietic recovery upon 5-fluorouracil-induced injury. Utilizing a bone marrow transplant model with a barcoded microRNA library, we screened for barcode abundance in peripheral blood of recipient mice before and after 5-fluorouracil treatment. Overexpression of screen-candidate miR-150 resulted in significantly slowed recovery rates across major blood lineages, with associated impairment of bone marrow clonogenic potential. Conversely, platelets and myeloid cells from miR-150 null marrow recovered faster after 5-fluorouracil treatment. Heterozygous knockout of c-myb, a conserved target of miR-150, partially phenocopied miR-150-forced expression. Our data highlight the role of microRNAs in controlling hematopoietic injury response and demonstrate the power of in vivo functional screens for studying microRNAs in normal tissue physiology.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nature immunology. 2008;9:839–845. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

