Homepeptide repeats: implications for protein structure, function and evolution
- PMID: 23084777
- PMCID: PMC5054710
- DOI: 10.1016/j.gpb.2012.04.001
Homepeptide repeats: implications for protein structure, function and evolution
Abstract
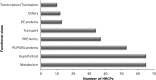
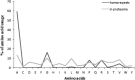
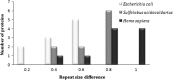
Analysis of protein sequences from Mycobacterium tuberculosis H37Rv (Mtb H37Rv) was performed to identify homopeptide repeat-containing proteins (HRCPs). Functional annotation of the HRCPs showed that they are preferentially involved in cellular metabolism. Furthermore, these homopeptide repeats might play some specific roles in protein-protein interaction. Repeat length differences among Bacteria, Archaea and Eukaryotes were calculated in order to identify the conservation of the repeats in these divergent kingdoms. From the results, it was evident that these repeats have a higher degree of conservation in Bacteria and Archaea than in Eukaryotes. In addition, there seems to be a direct correlation between the repeat length difference and the degree of divergence between the species. Our study supports the hypothesis that the presence of homopeptide repeats influences the rate of evolution of the protein sequences in which they are embedded. Thus, homopeptide repeat may have structural, functional and evolutionary implications on proteins.
Copyright © 2012. Published by Elsevier Ltd.
Figures


References
-
- Nakachi Y., Hayakawa T., Oota H., Sumiyama K., Wang L., Ueda S. Nucleotide compositional constraints on genomes generate alanine-, glcyine-, and proline-rich structures in transcription factors. Mol Biol Evol. 1997;14:1042–1049. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

