Host translation at the nexus of infection and immunity
- PMID: 23084916
- PMCID: PMC7104986
- DOI: 10.1016/j.chom.2012.09.006
Host translation at the nexus of infection and immunity
Abstract
By controlling gene expression at the level of mRNA translation, organisms temporally and spatially respond swiftly to an ever-changing array of environmental conditions. This capacity for rapid response is ideally suited for mobilizing host defenses and coordinating innate responses to infection. Not surprisingly, a growing list of pathogenic microbes target host mRNA translation for inhibition. Infection with bacteria, protozoa, viruses, and fungi has the capacity to interfere with ongoing host protein synthesis and thereby trigger and/or suppress powerful innate responses. This review discusses how diverse pathogens manipulate the host translation machinery and the impact of these interactions on infection biology and the immune response.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
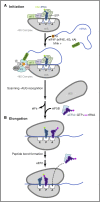

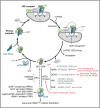
Similar articles
-
Mouse Norovirus Infection Arrests Host Cell Translation Uncoupled from the Stress Granule-PKR-eIF2α Axis.mBio. 2019 Jun 18;10(3):e00960-19. doi: 10.1128/mBio.00960-19. mBio. 2019. PMID: 31213553 Free PMC article.
-
Down the rabbit hole: Is necroptosis truly an innate response to infection?Cell Microbiol. 2017 Aug;19(8). doi: 10.1111/cmi.12750. Epub 2017 Jun 13. Cell Microbiol. 2017. PMID: 28476074 Review.
-
Influenza A virus-induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression.J Virol. 2014 Aug;88(15):8375-85. doi: 10.1128/JVI.00126-14. Epub 2014 May 14. J Virol. 2014. PMID: 24829357 Free PMC article.
-
HIV Exploits Antiviral Host Innate GCN2-ATF4 Signaling for Establishing Viral Replication Early in Infection.mBio. 2017 May 2;8(3):e01518-16. doi: 10.1128/mBio.01518-16. mBio. 2017. PMID: 28465428 Free PMC article.
-
The ubiquitin system: a critical regulator of innate immunity and pathogen-host interactions.Cell Mol Immunol. 2016 Sep;13(5):560-76. doi: 10.1038/cmi.2016.40. Epub 2016 Aug 15. Cell Mol Immunol. 2016. PMID: 27524111 Free PMC article. Review.
Cited by
-
Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals.Elife. 2016 May 31;5:e14228. doi: 10.7554/eLife.14228. Elife. 2016. PMID: 27240734 Free PMC article.
-
Virus-induced translational arrest through 4EBP1/2-dependent decay of 5'-TOP mRNAs restricts viral infection.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):E2920-9. doi: 10.1073/pnas.1418805112. Epub 2015 May 18. Proc Natl Acad Sci U S A. 2015. PMID: 26038567 Free PMC article.
-
Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens.Nat Rev Microbiol. 2013 Jun;11(6):365-9. doi: 10.1038/nrmicro3029. Epub 2013 May 13. Nat Rev Microbiol. 2013. PMID: 23669888 Review.
-
Repression of eEF2K transcription by NF-κB tunes translation elongation to inflammation and dsDNA-sensing.Proc Natl Acad Sci U S A. 2019 Nov 5;116(45):22583-22590. doi: 10.1073/pnas.1909143116. Epub 2019 Oct 21. Proc Natl Acad Sci U S A. 2019. PMID: 31636182 Free PMC article.
-
A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication.Genes Dev. 2013 Aug 15;27(16):1809-20. doi: 10.1101/gad.221341.113. Genes Dev. 2013. PMID: 23964095 Free PMC article.
References
-
- Axten J.M., Medina J.R., Feng Y., Shu A., Romeril S.P., Grant S.W., Li W.H., Heerding D.A., Minthorn E., Mencken T. Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK) J. Med. Chem. 2012;55:7193–7207. - PubMed
-
- Bauman Y., Nachmani D., Vitenshtein A., Tsukerman P., Drayman N., Stern-Ginossar N., Lankry D., Gruda R., Mandelboim O. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9:93–102. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

